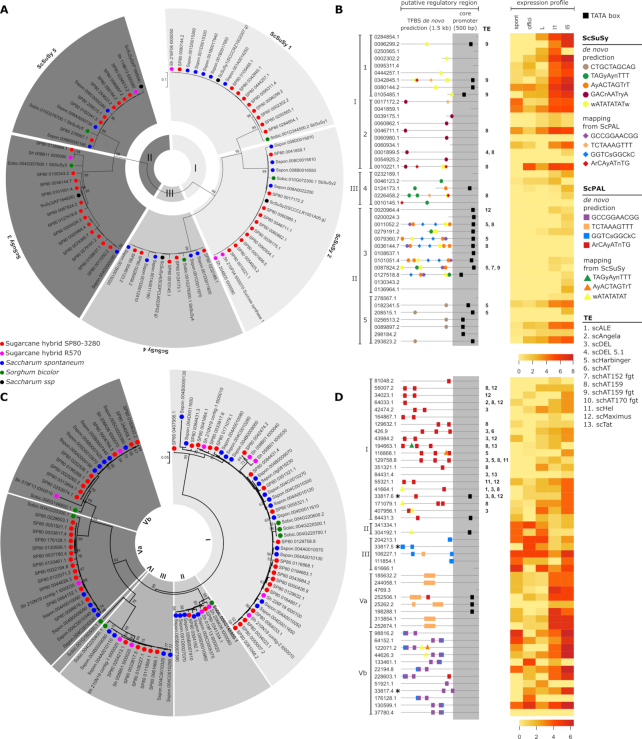
Figure 4:
Phylogeny, putative regulatory regions, and expression of sucrose synthase (SuSy) and phenylalanine-ammonia lyase (PAL) gene family. Phylogenetic analysis of (A) SuSy and (C) PAL genes from SP80-3280, R570, S. spontaneum, and sorghum. SuSy sequences from Saccharum ssp. [36] were also included. For both SuSy and PAL, nucleotide sequences (CDS) were aligned with CLUSTALW [37] software in MEGA 7.0 [38] and maximum likelihood trees were constructed with 1,000 bootstraps. Core promoter analysis (gray columns in B and D) using TSSPlant [39] suggests ScSuSy2 (B) and most ScPAL (D) as TATA-less (absence of black squares). Transcription factor binding site (TFBS) prediction (colored symbols in B and D) using MEME [40] and MotifSampler [41] suggests specific motif for each group (ScSuSy1, ScSuSy2, and ScSuSy5 and PAL I, PAL III, PAL Va, and PAL Vb). The three SP80-3280 PAL genes marked with an asterisk in D are present in the same contig. Transposable elements (TEs) were identified within 10 kb upstream from the gene (B and D). Heat map analysis of RNA-Seq data [29] (expression profile in B and D) shows more pronounced expression in SP80-3280 internodes (I1 and I5) of ScSuSy1, ScSuSy2, ScSuSy5, and PAL from group V. RNA-Seq of leaf tissues (L) indicates more pronounced expression of ScPAL from groups II and III. ScSuSy3 presents high numbers of TFBS and TE and low expression in all samples.hylogeny, putative regulatory regions, and expression of sucrose synthase (SuSy) and phenylalanine-ammonia lyase (PAL) gene family. Phylogenetic analysis of (A) SuSy and (C) PAL genes from SP80-3280, R570, S. spontaneum, and sorghum. SuSy sequences from Saccharum ssp. [36] were also included. For both SuSy and PAL, nucleotide sequences (CDS) were aligned with CLUSTALW [37] software in MEGA 7.0 [38] and maximum likelihood trees were constructed with 1,000 bootstraps. Core promoter analysis (gray columns in B and D) using TSSPlant [39] suggests ScSuSy2 (B) and most ScPAL (D) as TATA-less (absence of black squares). Transcription factor binding site (TFBS) prediction (colored symbols in B and D) using MEME [40] and MotifSampler [41] suggests specific motif for each group (ScSuSy1, ScSuSy2, and ScSuSy5 and PAL I, PAL III, PAL Va, and PAL Vb). The three SP80-3280 PAL genes marked with an asterisk in D are present in the same contig. Transposable elements (TEs) were identified within 10 kb upstream from the gene (B and D). Heat map analysis of RNA-Seq data [29] (expression profile in B and D) shows more pronounced expression in SP80-3280 internodes (I1 and I5) of ScSuSy1, ScSuSy2, ScSuSy5, and PAL from group V. RNA-Seq of leaf tissues (L) indicates more pronounced expression of ScPAL from groups II and III. ScSuSy3 presents high numbers of TFBS and TE and low expression in all samples.