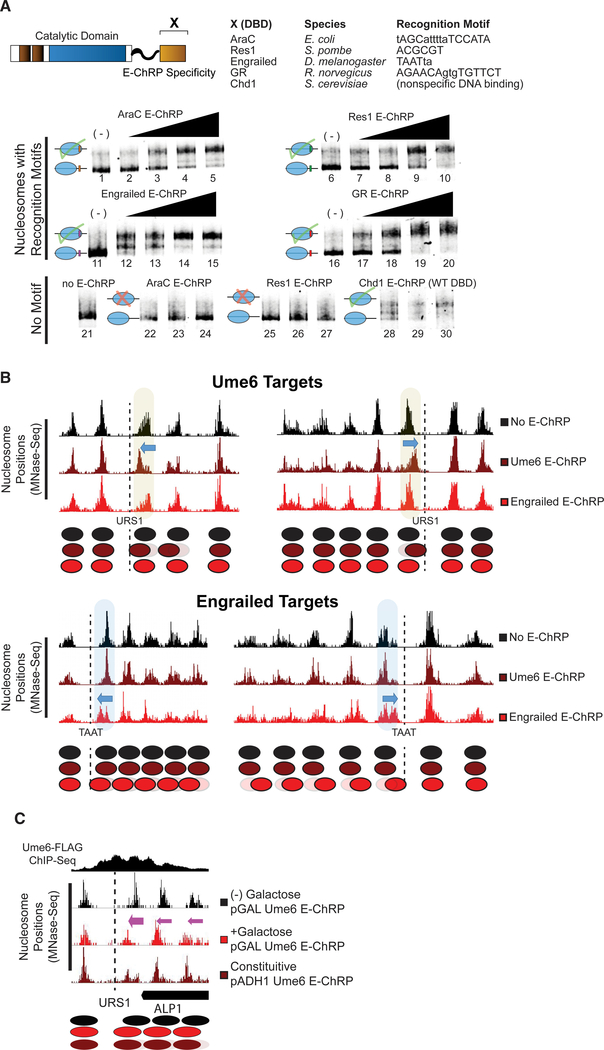
Figure 2. E-ChRPs with Distinct TF DBDs Specifically Position Target Nucleosomes In Vitro and In Vivo.
(A) Nucleosome sliding assay demonstrating functionality of Increasing concentrations of E-ChRPs containing AraC DBD (lanes 1–5 and 22–24), Res1 DBD (lanes 6–10 and 25–27), engrailed DBD (lanes 11–15), glucocorticoid receptor DBD (lanes 16–20), or Chd1 endogenous DBD (lanes 28–30) in vitro. Nucleosomes in lanes 1–20 possess recognition motifs in extranucleosomal DNA for the respective E-ChRP (Ades and Sauer, 1994; Alroy and Freedman, 1992; Anderson et al., 1995; Ayté et al., 1995; Khan et al., 2018; Niland et al., 1996), while lanes 21–30 have no recognition motif. Lower electrophoretic mobility indicates repositioning of nucleosomes away from their end positions.
(B) Yeast genomic nucleosome dyad positions are shown at representative Ume6 targets (URS1, top) or engrailed targets (TAAT, bottom) in the presence or absence of Ume6 E-ChRP or engrailed E-ChRP. Motif-proximal nucleosomes are highlighted next to indicated motifs, with blue arrows showing direction of nucleosome movement. Cartoon representations of nucleosome positions are provided for each locus.
(C) Nucleosome dyad signal at a representative locus in yeast demonstrating positioning of nucleosomes toward recruitment motif (dashed line) by galactose-inducible Ume6 E-ChRP or a constitutively expressed E-ChRP under the ADH1 promoter.
See also Figure S1.