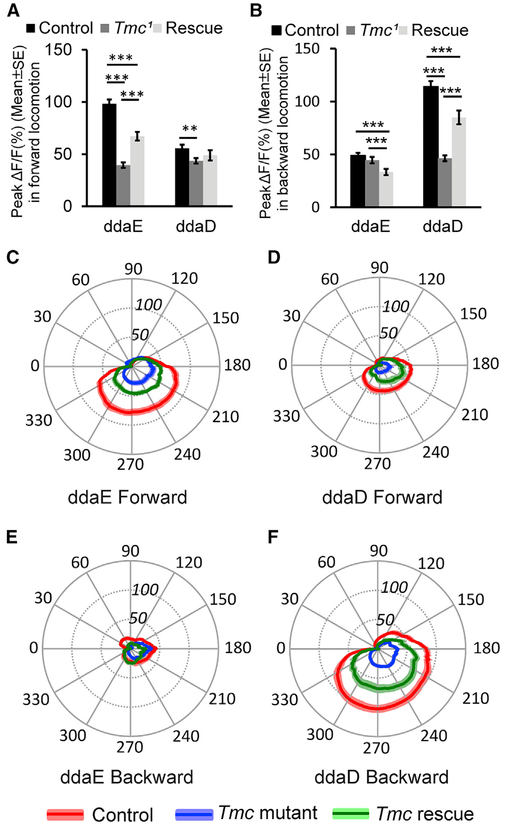
Figure 7. Tmc Is Required for Idiothetic Activation of ddaE and ddaD in Larval Forward and Backward Locomotion.
(A) Comparison of peak ΔF/F (percentage) in ddaE and ddaD neurons in Tmc wild-type control (n = 54 for ddaE and n = 51 for ddaD), mutant (n = 36 for ddaE and n = 43 for ddaD), and rescue (n = 19 for both ddaE and ddaD) larvae in larval forward locomotion.
(B) Comparison of peak ΔF/F (%) in ddaE and ddaD in Tmc wild-type control (n = 36 for ddaE and n = 38 for ddaD), mutant (n = 35), and rescue (n = 20) larvae in larval backward locomotion.
(C) Comparison of mean ΔF/F (%) in ddaE of Tmc wild-type control (n = 25), mutant (n = 17), and rescue (n = 16) larvae in larval forward locomotion during the segmental contraction cycle.
(D) Comparison of mean ΔF/F (%) in ddaD of Tmc wild-type control (n = 25), mutant (n = 17), and rescue (n = 15) larvae in larval forward locomotion in the segmental contraction cycle.
(E) Comparison of mean ΔF/F (%) in ddaE of Tmc wild-type control (n = 17), mutant (n = 26), and rescue (n = 18) larvae in larval backward locomotion in a muscle contraction cycle.
(F) Comparison of mean ΔF/F (%) in ddaD of Tmc wild-type control (n = 21), mutant (n = 26), and rescue (n = 18) larvae in larval backward locomotion in a muscle contraction cycle.
Genotypes are as follows: Tmc wild-type control is w; UAS-GCaMP6f;2-21- GAL4. Tmc mutantis w; UAS-GCaMP6f;2-21-GAL4 Tmc1. Tmc rescue is w;UAS-GCaMP6f UAS-Tmc; 2-21-GAL4 Tmc1. ** p < 0.01, ***p < 0.001, Student’s t test. Error bars in (A) and (B) indicate the SEM. The position of the radial axis in (C)-(F) represents DF/F (percentage). Solid colored lines represent the mean values, and colored shading denotes the SEM. n indicates the number of neurons examined. Neuronal activities in forward movements were recorded from nine, ten, and seven animals for Tmc wild-type control, Tmc mutant, and Tmc rescue, respectively. Neuronal activities in backward movements were recorded from eleven, eleven, and nine animals for Tmc wildtype control, Tmc mutant, and Tmc rescue, respectively. See also Figures S2 and S5 and Video S7