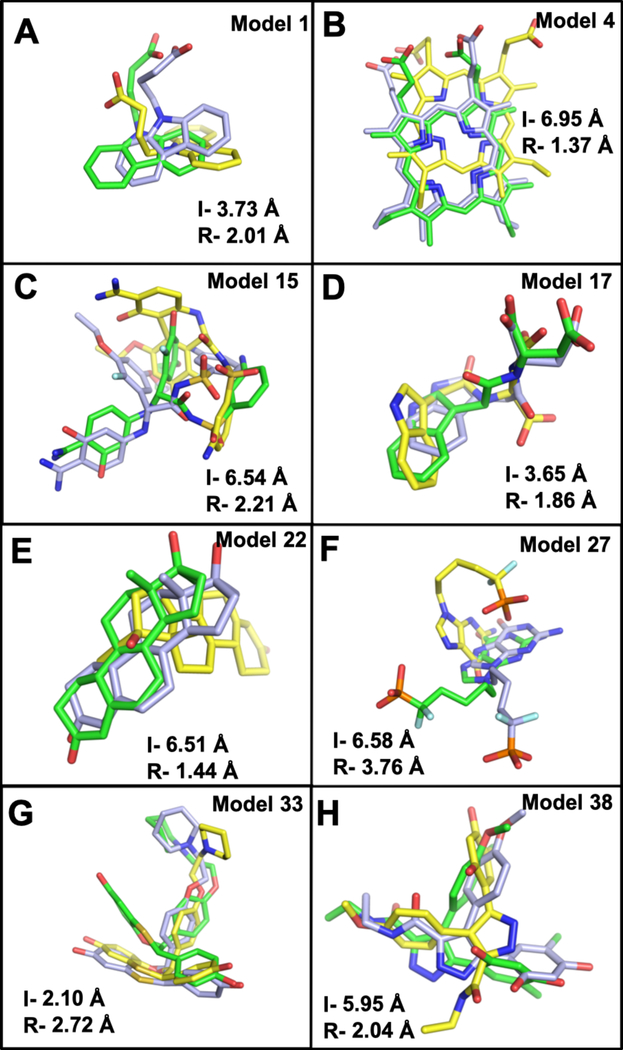
Figure 5.
Ligand binding modes obtained from AutoDock Vina are compared to the crystal structures (light blue). Ligand poses obtained from docking to the initial unrefined structures (yellow) and docking to the refined structure (green). I- is the initial unrefined structure ligand RMSD, and R- refers to refined structure ligand RMSD. Two representatives from each group are shown, (A-B) group 1: model 4 – deoxyhemoglobin (PDB 1g9v) and model 1- FABP, adipocyte (PDB 1tow). (C-D) Group 2: model 15-serine protease factor Vila (PDB 1ygc) and model 17- tryptophan synthase (PDB 1k3u). (E-F) Group 3: model 22- progesterone receptor (PDB 1sqn) and model 27- purine nucleoside phosphorylase (PDB 1v48). (G-H) Group 4: model 33- estrogen receptor (PDB 1sj0) and model 38- HSP (PDB 2bsm).