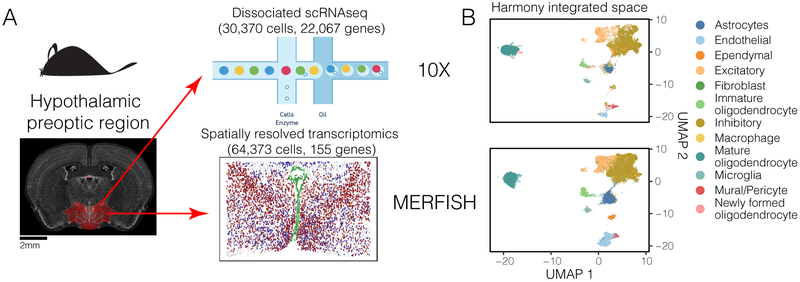
Figure 6.
Harmony integrates spatially resolved transcriptomic with dissociated scRNAseq datasets. (A) Cells from the hypothalamic preoptic region of mouse brain were assayed in parallel with two technologies. The full transcriptome of dissociated cells was profiled with 10X. 155 genes were profiled in-situ on intact tissue with MERFISH. (B) Harmony integrated cells from the two modalities into a shared embedding, correctly merging the 12 previously identified cell types. (C) Satb1 expression (blue), unmeasured in the MERFISH dataset, was inferred and predicted to be spatially autocorrelated in inhibitory neurons. Satb1 expression was highest in anterior slices and diminished in slices that contained ventricle-lining Ependymal cells (green). (D) Matched images from an independent in-situ hybridization experiment measuring Satb1 expression from the Allen Brain Atlas. Satb1 expression (blue) is co-localized in similar regions of the slices and diminishes with the appearance of ventricle structures (green).