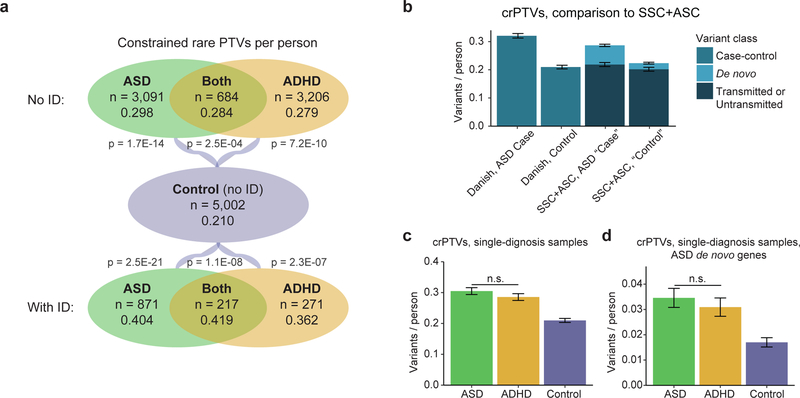
Figure 1: Rates of constrained rare protein-truncating variants (crPTVs).
a) Mean rates of crPTVs across phenotypes, with and without intellectual disability (ID). “Constrained” denotes genes with pLI (probability of being loss-of-function intolerant) values at least 0.9. “Rare” denotes variants with an allele count of no greater than 5 across the 13,342 Danish samples analyzed in this study and the 44,779 non-Finnish Europeans in the non-psychiatric exome subset of gnomAD (58,121 total individuals). P values shown are for comparison to controls. Differences between case categories without ID are not significant (p = 0.49 for ASD vs ASD+ADHD; p = 0.91 for ADHD vs ASD+ADHD; p = 0.14 for ASD vs ADHD), nor are differences between case categories with ID significant (p = 0.59 for ASD vs ASD+ADHD; p = 0.60 for ADHD vs ASD+ADHD; p = 0.58 for ASD vs ADHD). b) Mean rates of crPTVs in Danish case-control data compared to crPTVs in Simons Simplex Collection (SSC) and Autism Sequencing Consortium (ASC) family-based data. From SSC+ASC data14, we constructed ASD “cases” using de novo variants from affected probands (n = 3,982) and transmitted variants from parents of probands (n = 4,319), and we constructed “controls” using de novo variants from unaffected children (n = 2,078) and untransmitted variants from parents of probands (n = 4,319). SSC+ASC variants were counted as “rare” if they had an allele count ≤ 5 across the SSC+ASC data and non-Finnish Europeans from the non-psychiatric exome subset of gnomAD. Danish data is from all individuals with an ASD diagnosis (including comorbid ADHD and/or intellectual disability, n = 4,863) and controls (n = 5,002), and “rare” is defined as in part a. c-d: Mean rates of crPTVs in ASD cases (n = 2,430) and ADHD cases (n = 2,360) with only a single diagnosis (i.e. no comorbid ASD+ADHD samples, no intellectual disability diagnosis, and no diagnoses of schizophrenia, bipolar disorder, affective disorder, or anorexia). “Rare” is defined as in part a, and the same controls (n = 5,002) are used. c) Rates in all constrained genes. ASD and ADHD rates are not significantly different from each other (p = 0.21), while both are significantly different from controls (OR = 1.46 for ASD based on 741 crPTVs, p = 1.12E-14; OR = 1.37 for ADHD based on 674 crPTVs, p = 2.26E-10; 1,049 crPTVs in controls). d) Rates in the 212 constrained genes with a published rare de novo PTV in ASD (“ASD de novo genes”)14. ASD and ADHD rates are again not significantly different from each other (p = 0.38), while both are significantly different from controls (OR = 2.19 for ASD based on 84 crPTVs, p = 5.39E-07; OR = 1.87 for ADHD based on 73 crPTVs, p = 1.40E-04; 85 crPTVs in controls). For a-d, all p values are by logistic regression (Methods), and all error bars are Poisson standard error. OR = odds ratio.