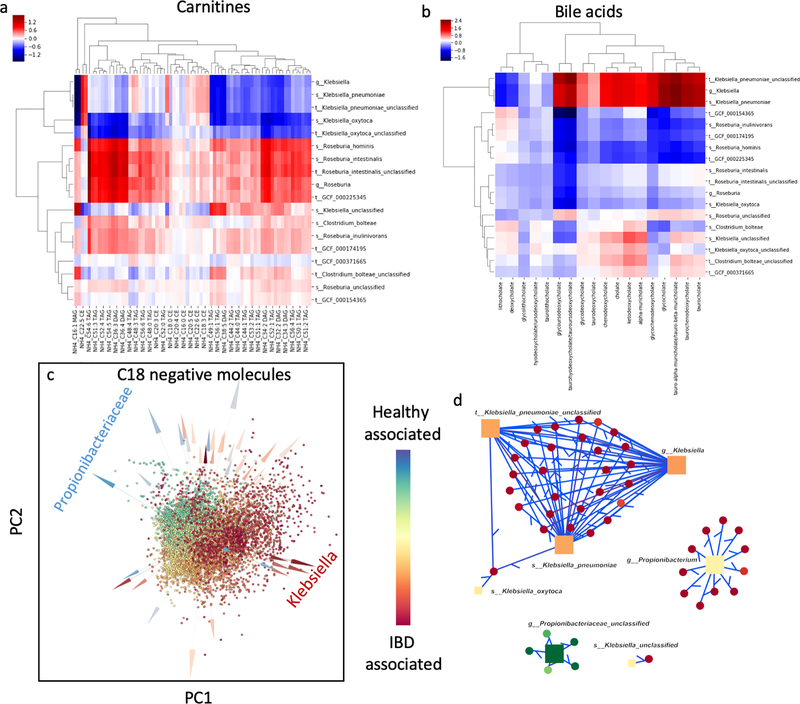
Figure 6:
Microbe-metabolite interactions of the human microbiome in association with IBD samples [29]. (a) Heatmap visualization of the inferred conditional probabilities for various bile acids given the presence of Klebsiella, Roseburia and Clostridium bolteae. (b) Heatmap visualization of the inferred conditional probabilities for the carnitines given the presence of Klebsiella, Roseburia, and Clostridium bolteae. (c) Multiomics biplot of the microbe-metabolite interactions learned from metagenomics profiles and C18 negative ion mode LC-MS. Microbes (arrows) and metabolites (spheres) are colored according to their differentials estimated from multinomial regression. Klebsiella spp. appears to be strongly associated with IBD, while Propionibacterium spp. has strong negative association. (d) Network of the top 300 edges where only the edges that contain Klebsiella and Propionibacteriaceae are visualized.