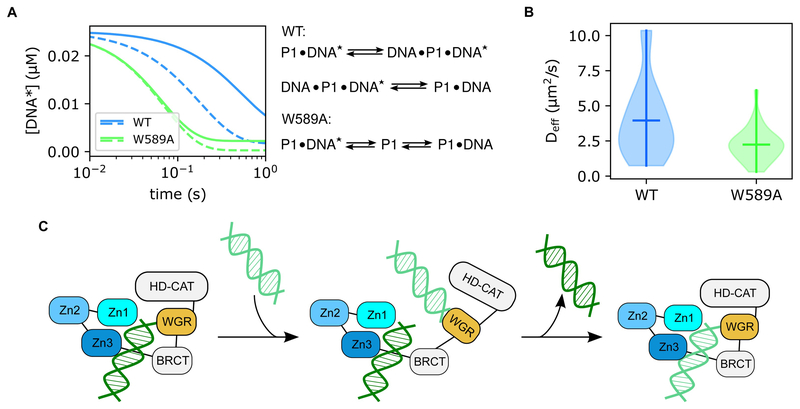
Figure 3.
The “monkey bar” mechanism of PARP1 mobility. (A) Stopped-flow kinetics models show that wild-type PARP1 (blue) releases pre-bound DNA fragments ([DNA*]) at a significantly slower rate than the W589A mutant (green). The W589A mutant showed no difference in off-rate observation between low A Timeline of Microirradiation-based Studies(0.4 μM, solid line) and high (4 μM, dashed line) concentrations of competitor DNA, but wild-type PARP1 displays faster release of the initially bound strand when exposed to a higher concentration of competitor DNA. In addition, fitting of W589A kinetics were adequately described by a two-state equation, whereas wild-type data required formation of an intermediate state. (B) Violin plots of modeled Deff values from Q-FADD analyses on wild-type (blue) and W589A (green) microirradiation data. These models show that wild-type PARP1 diffuses significantly more quickly to DNA lesions than the W589A mutant (p-value = 0.0094). (C) Graphical representation of the “monkey bar” mechanism derived from the combination of these data. The WGR domain of PARP1 releases from the currently bound DNA strand and binds to a neighboring strand of DNA, then the original strand of DNA is released and the complex collapses around the newly bound DNA fragment. Figures were adapted from our published (free-access) data [18].