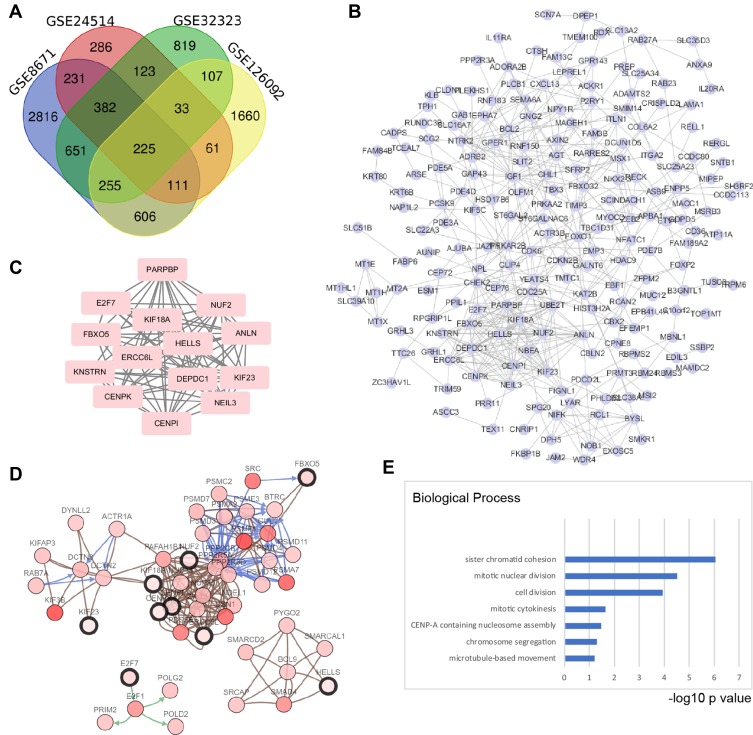
Figure 1.
Screening hub genes in colorectal cancer by bioinformatics methods based on GEO data sets. (A) Four data sets were selected: GSE8671, GSE24514, GSE32323, and GSE126092. For screening the DEGs, the GEO2R tool was used, the cut-off value for adjusted p-value was 0.01, and the fold change (Log2) was ±0.75. A total of 225 overlap genes were found in the four data sets. (B) The protein-protein interaction network (PPI) was predicted by the STRING online tool, and then the interactions among the 225 genes were reconstructed by Cytoscape (Version 3.7.1). (C) Hub genes were screened by the Cytoscape plug-in APP MCODE (Version 1.4.1); an MCODE score > 10 was selected, which resulted in 14 hub genes. (D) These hub genes were correlated with TCGA data by cBioPortal (TCGA, colorectal adenocarcinoma, provisional), which were used to reconstruct the coexpression network; five functional clusters were found. (E) Gene Ontology analysis of hub genes was performed by DAVID online tool. The top 4 biological processes were sister chromatid cohesion, mitotic nuclear division, cell division and mitotic cytokinesis. The X axis shows the rank by the -log10 p value of each enriched process.