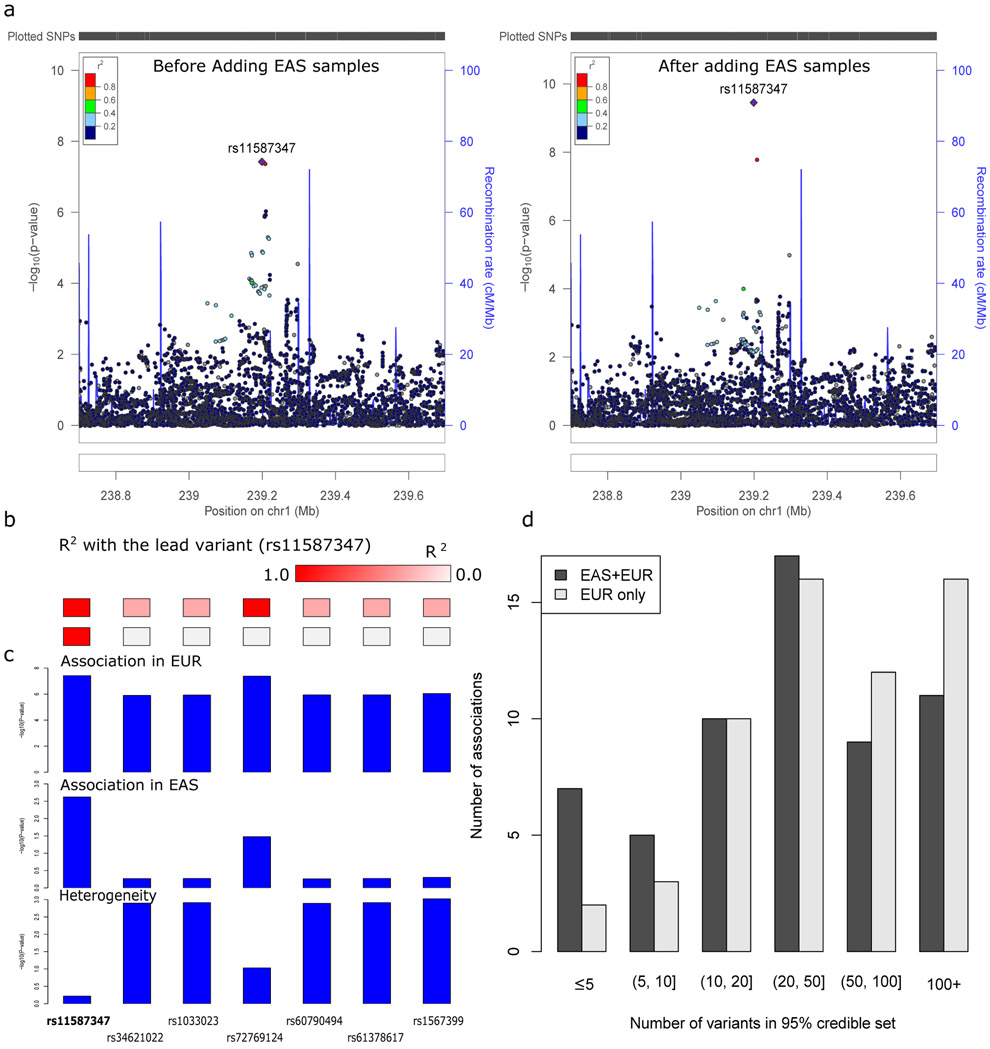
Figure 3 ∣. Trans-ethnic fine-mapping improves resolution.
a, An association was mapped to a single variant (rs11587347) after adding EAS samples and using the trans-ancestry fine-mapping approach. Regional association plots were generated using http://locuszoom.org/ and LD from 1000 Genomes Project Phase 3 EUR subjects. b, LD with the lead variant (rs11587347). c, The lead variant (rs11587347) has strong association significance in both populations and low heterogeneity across populations. a-c, n (EAS stage 1) = 13,305 cases, 16,244 controls; n (EUR PGC2) = 33,640 cases, 43,456 controls. d, Number of variants in the 95% credible set using the trans-ancestry (EAS+EUR) and published fine-mapping approaches (EUR only).