Abstract
The recently-discovered single-span transmembrane proteins endoregulin (ELN), dwarf open reading frame (DWORF), myoregulin (MLN), and another-regulin (ALN) are reported to bind to the SERCA calcium pump in a manner similar to that of known regulators of SERCA activity, phospholamban (PLB) and sarcolipin (SLN). To determine how micropeptide assembly into oligomers affects the availability of the micropeptide to bind to SERCA in a regulatory complex, we used co-immunoprecipitation and fluorescence resonance energy transfer (FRET) to quantify micropeptide oligomerization and SERCA-binding. All micropeptides formed avid homo-oligomers with high-order stoichiometry (n>2 protomers per homo-oligomer), but it was the monomeric form of all micropeptides that interacted with SERCA. In view of these two alternative binding interactions, we evaluated the possibility that oligomerization occurs at the expense of SERCA-binding. However, even the most avidly oligomeric micropeptide species still showed robust FRET with SERCA, and there was a surprising positive correlation between oligomerization affinity and SERCA-binding. This comparison of micropeptide family members suggests that the same structural determinants that support oligomerization are also important for binding to SERCA. Moreover, the unique oligomerization/SERCA-binding profile of DWORF is in harmony with its distinct role as a PLB-competing SERCA activator, in contrast to the inhibitory function of the other SERCA-binding micropeptides.
Graphical abstract
Introduction:
The sarco(endo)plasmic reticulum calcium ATPase is the principal transport mechanism for sequestration of calcium (Ca) in the endoplasmic reticulum (ER). These intracellular Ca stores are the foundation of signals that coordinate diverse cellular functions. For example, in muscle cells, sarcoplasmic reticulum (SR) Ca release and reuptake are the signals for contraction and relaxation. Different tissues vary in their Ca uptake requirements, and differential expression of distinct SERCA isoforms provides some tissue-specific specialization of Ca handling. In addition, the kinetics and Ca sensitivity of transport are modulated by single-span proteins, regulatory “micropeptides”, which interact with the transmembrane domain of SERCA. These include the well-known SERCA regulatory partners phospholamban (PLB)1 and sarcolipin (SLN)2, which are inhibitors of SERCA in cardiac and skeletal muscle. New additions to this family include dwarf open reading frame (DWORF), myoregulin (MLN), endoregulin (ELN), expressed in cardiac muscle, skeletal muscle, and endo/epithelial tissues, respectively3–5, and another-regulin (ALN), expressed ubiquitously. With these newly identified micropeptides, SERCA regulatory complexity is beginning to resemble that of a related ion transporter, the Na+/K+-ATPase (NKA), which has many tissue-specific, species-specific regulatory peptides6, 7. The differential expression of diverse micropeptides4 may precisely tune the function of ion transporter isoforms to the needs of muscle and non-muscle cells. Thus, peptide-transporter regulatory interactions are relevant to a variety of disciplines including cardiology, neuroscience, and endocrinology.
There is a great deal of excitement about these new modes of SERCA regulation as they may represent new opportunities for therapeutic intervention, but at this early stage the structure/function mechanisms of micropeptides are still poorly understood. One key functional determinant must be the stability of the regulatory complex of the micropeptide with the target transporter. Different micropeptides co-expressed in the same tissue may compete for SERCA5, 8, with a net effect depending on the relative expression and binding affinity of each species. Despite decades of study of the prototypical micropeptide, PLB, it has been difficult to determine precisely the extent to which SERCA is regulated by this interaction in vivo. As reviewed by Ceholski et al.9, there are a wide range of estimates of PLB:SERCA ratios reported in the literature, from 1:5 to 4:110–14. We think that the larger PLB:SERCA ratios are more likely to be correct, but even a molar excess of PLB does not necessarily reflect stoichiometric regulation of SERCA. Overexpression of PLB in a transgenic mouse model resulted in increased SERCA functional regulation, suggesting some SERCA was not regulated at native expression levels of PLB15. Nevertheless, this apparent substoichiometric regulation is still physiologically significant, and relief of inhibition results in robust enhancement of calcium handling to support increased cardiac function during exercise.
One reason for substoichiometric regulation of SERCA in spite of molar excess of PLB is that the micropeptide forms pentamers that do not regulate the transporter. More generally, oligomerization of micropeptides may indirectly modulate transporter function by altering micropeptide availability, thereby tuning overall functional potency. This concept of linked equilibria of SERCA-binding and oligomerization was developed from PLB mutagenesis experiments in which destabilization of the PLB pentamer increased SERCA binding and inhibition16–19. In a reciprocal experiment, increasing the proportion of SERCA resulted in decreased PLB oligomerization20, suggesting that SERCA sequestered monomeric PLB. Also, phosphorylation21–23 or oxidative crosslinking24, 25 of PLB increased oligomerization and decreased SERCA-binding. However, significant exceptions to this model exist in the form of PLB variants that are strong oligomers and still bind avidly to SERCA26, 27, or weakly oligomeric PLB variants that bind weakly to SERCA28.
Here we investigated the interplay of oligomerization and SERCA-binding across the micropeptide family. To define the relative affinities of the micropeptide regulatory complexes, we used FRET to measure the dissociation constant of the micropeptide homo-oligomers (KD1) and the micropeptide-SERCA complexes (KD2). We focused on the cardiac isoform, SERCA2a, as this transporter is of particular interest as a possible point of intervention against heart disease. Besides its central role in normal cardiac physiology, the SERCA pump has been implicated in the impaired calcium handling that underlies many types of heart failure29, 30. While the etiology of heart failure is complex, mutations in calcium-handling proteins cause cardiomyopathy31–34, and restoring calcium homeostasis has improved function and survival in animal models of heart failure35, 36. This has focused considerable attention on SERCA as a therapeutic target30, 35–37. Though SERCA gene delivery has yielded only modest gains in SERCA expression in patients38, efforts to deliver exogenous SERCA to the failing heart are continuing. In addition, there is interest in parallel approaches based on improving the function of the endogenous SERCA remaining in the diseased myocardium. For this, we need a better understanding of fundamental functional determinants.
Results:
Micropeptide-SERCA Regulatory Interactions
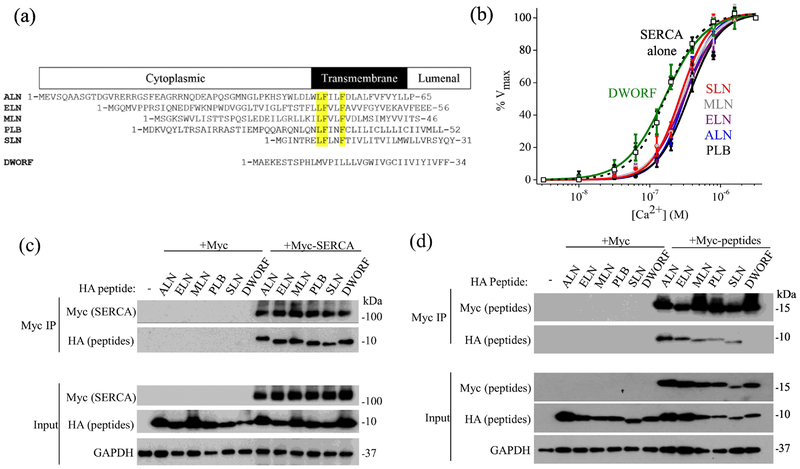
A comparison of the sequences of micropeptides is shown in Fig. 1A, with key conserved residues highlighted. Previous studies have suggested a direct physical interaction between SERCA and micropeptides3, 4, 8, 21, 39. Here we observed comparable efficiencies of coimmunoprecipitation of HA-tagged micropeptides PLB, SLN, DWORF, ALN, ELN, and MLN with Myc-SERCA (Fig. 1B). Since homo-oligomerization may affect micropeptide availability for binding to SERCA, we also tested whether each HA-tagged micropeptide co-immunoprecipitated with the same micropeptide tagged with Myc (Fig. 1C). With the exception of DWORF, all HA-micropeptides co-immunoprecipitated with the corresponding Myc-micropeptide partner, suggesting the micropeptides physically associate in homo-oligomeric complexes. To assess SERCA regulation by micropeptides, we performed oxalate-supported 45Ca-uptake measurements and determined all micropeptides decreased the apparent affinity of SERCA for Ca, with the exception of DWORF (Fig. 1D). DWORF yielded a Ca-dependence of transport that was not distinguishable from control (SERCA alone). This observation is consistent with the model in which DWORF binds SERCA but does not inhibit the pump5, 8, enhancing Ca uptake by displacing PLB. Values of measured KCa for Ca uptake are provided in Table 1.
Fig. 1.
Regulation of SERCA by micropeptides. A) Sequence alignment of micropeptides, with key conserved residues highlighted. B) Compared to control (SERCA alone, empty squares, black dotted line), the apparent Ca affinity of transport activity was decreased when SERCA was co-expressed with PLB (black circles, black solid line), SLN (red), ALN (blue), ELN (purple), and MLN (gray). The Ca sensitivity of SERCA co-expressed with DWORF (green) was not significantly decreased. C) and D) Pull down of Myc-SERCA (C) or Myc-micropeptide (D) resulted in coimmunoprecipitation of HA-tagged micropeptide, suggesting a physical interaction in transfected HEK cells.
Table 1.
Results of acceptor sensititzation FRET measurements. Best estimates of intra-oligomeric FRET distances are shown in bold fon.
| KCa (nM) | ||||||
|---|---|---|---|---|---|---|
| ALN | 3.9 ± 0.8 | 56.9 ± 8.6 | 47.2 | ALN | 472.0 ± 76.6 | |
| ELN | 4.7 ± 0.5 | 63.5 ± 8.1 | 45.0 | ELN | 471.8 ± 28.5 | |
| PLB | 1.5 ± 0.4 | 26.7 ± 3.6 | 58.5 | PLB | 532.7 ± 86.7 | |
| DWORF | 1.6 ± 0.7 | 34.3 ± 7.7 | 54.4 | DWORF | 208.8 ± 32.0 | |
| SLN | 2.9 ± 0.8 | 39.0 ± 7.3 | 51.2 | SLN | 384.0 ± 24.5 | |
| MLN | 2.0 ± 0.1 | 20.0 ± 0.5 | 61.8 | MLN | 436.3 ± 32.8 | |
| Control | 218.0 ± 5.1 | |||||
| Distance (Å) | ||||||
| Pentamer | ||||||
| ALN | 2.8± 0.5 | 52.5 ± 3.1 | 48.2 | 56.2 | 57.0 | 57.0 |
| ELN | 3.1 ± 0.3 | 54.8 ± 3.9 | 46.8 | 55.0 | 55.9 | 55.8 |
| PLB | 0.2 ± 0.0 | 45.6 ± 2.8 | 49.8 | 58.3 | 58.8 | 58.7 |
| DWORF | 1.6 ± 1.6 | 32.2 ± 6.1 | 55.4 | 63.7 | 64.5 | 64.4 |
| SLN | 1.7 ± 0.2 | 64.7 ± 8.4 | 44.5 | 52.7 | 53.6 | 53.5 |
| MLN | 1.4 ± 0.5 | 57.1 ± 3.6 | 45.3 | 53.8 | 54.7 | 54.6 |
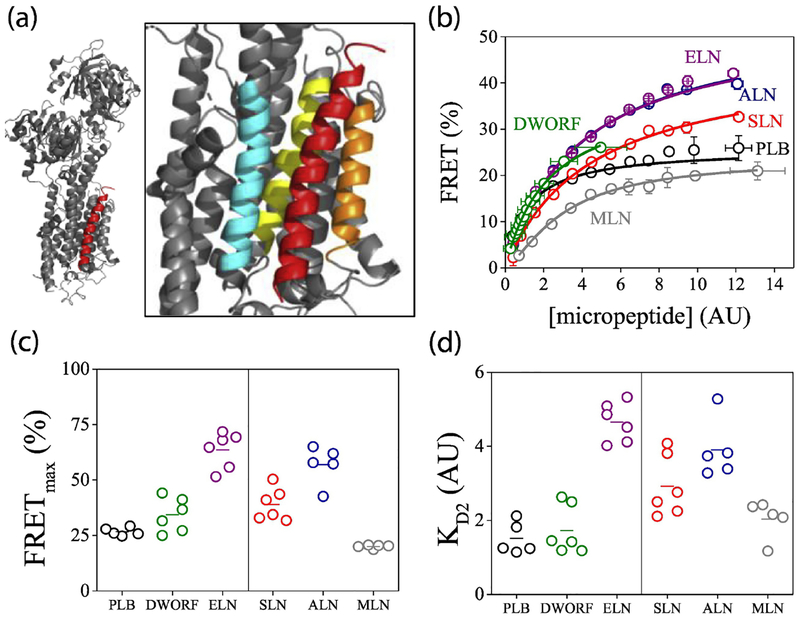
Fig. 2A shows the x-ray co-crystal structure of the SERCA-PLB regulatory complex40, revealing the site of interaction of the PLB transmembrane domain (red) with key transmembrane helices of the Ca transporter (grey). The enlarged inset shows that this canonical binding cleft is composed of SERCA transmembrane helices 2 (cyan), 6 (yellow), and 9 (orange), and other micropeptides are presumed to interact at the same location. Additional alternative binding sites have been proposed for phospholamban41, 42, but the occupation of these hypothetical alternative sites and their functional significance is unknown.
Fig. 2.
Quantification of the interaction of SERCA2a with micropeptides by acceptor sensitization. A) The structure of the regulatory complex of SERCA (gray) with the prototypical micropeptide PLB (red). Inset: enlarged view highlighting SERCA TM helices 2 (cyan), 6 (yellow), and 9 (orange). B) FRET from donor-labeled SERCA to acceptor-labeled micropeptides increased with increasing protein expression. Data are pooled from 2–6k cells per experiment. C) Fits of FRET curves with hyperbolic (left) or Hill (right) functions yielded estimates of maximal FRET (FRETmax), taken as the intrinsic FRET of SERCA-micropeptide regulatory complexes. D) Apparent dissociation constants (KD2) of SERCA-micropeptide complexes determined from hyperbolic (left) or Hill (right) function fitting.
Fluorescence resonance energy transfer (FRET) was used to generate “binding curves” to evaluate the affinity of SERCA-micropeptide complexes in the environment of the biological membrane in live cells17, 23, 43–47. This assay also provides insight into the overall architecture of the complexes by estimation of the distance from a FRET donor (fused to SERCA N-terminus) to a FRET acceptor (fused to a micropeptide N-terminus). Transient transfection of HEK cells resulted in a wide range of protein expression levels, as quantified from the YFP acceptor-labeled micropeptide fluorescence intensity. Cell-by-cell analysis of acceptor sensitization FRET with automated fluorescence microscopy43 revealed that FRET increased with protein expression up to a maximum. For clarity, data from cells with similar protein expression are pooled (Fig. 2B); examples of unpooled measurements of individual cells are provided in Supplemental Fig. 1. As previously described, the relationship between SERCA-PLB FRET and micropeptide expression (taken as an index of protein concentration) was well-described by a hyperbolic fit (Fig. 2B, black) of the form FRET=(FRETmax)([micropeptide])/(Kd+[micropeptide])17, 41, yielding the parameters of maximum FRET (FRETmax) and apparent dissociation constant (KD2). As a function of protein expression, FRET from SERCA to DWORF (Fig. 2B, green) and ELN (Fig. 2B, purple) also fit well with a hyperbola. However, the binding curves for SLN, ALN, and especially MLN were notable for apparent positive cooperativity, and the data were poorly described by a simple hyperbola. Therefore, these data were fit with a Hill function of the form FRET=(FRETmax)([micropeptide]n)/(Kdn +[micropeptide]n) where n is the Hill coefficient. SLN, ALN, and MLN binding curves yielded respective Hill coefficients of 1.19 ± 0.13, 1.14 ± 0.18, and 1.9 ± 0.13 (mean ± SD). The possible significance of this apparent positive cooperativity is discussed below. Fitting binding curves of other micropeptides with Hill functions did not significantly improve the quality of the fit or yield such Hill coefficients greater than 1.
Fig. 2C compares FRETmax values obtained by hyperbolic fits (left) or Hill fits (right) for 5–6 independent experiments with 2000–6000 cells per experiment. These values are taken as the intrinsic FRET of the SERCA-micropeptide complex, and the donor-acceptor separation distances they imply are provided in Table 1. Measurements were validated by acceptor sensitization FRET experiments with donor-acceptor fusion constructs of known FRET efficiency48. Apparent fluorescent probe separation distances ranged from 62 Å for MLN down to 45 Å for ELN, values that are broadly compatible with the expected distances between the N-terminal fusion site on SERCA and a micropeptide bound in the canonical binding site near SERCA helix M6 (Fig. 2A).
The relative affinities of the micropeptides for SERCA are compared in Fig. 2D, with KD2 values obtained from the hyperbolic (left) or Hill fits (right). The apparent KD2 of PLB was the lowest of the micropeptides tested here, suggesting that this prototypical SERCA regulator has the highest affinity for the pump. DWORF also showed an avid interaction with SERCA, consistent with its proposed role as a competitor for PLB. Overall, the relative affinities of micropeptides for SERCA were similar, with apparent KD2 values rank ordered as follows: PLB=DWORF= MLN<SLN<ALN=ELN.
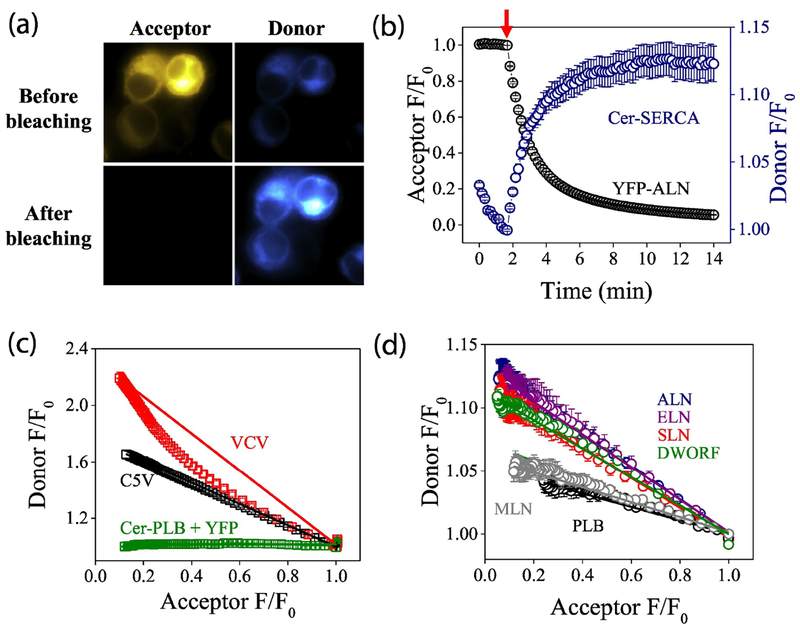
Micropeptide-SERCA complex stoichiometry quantified by progressive acceptor photobleaching
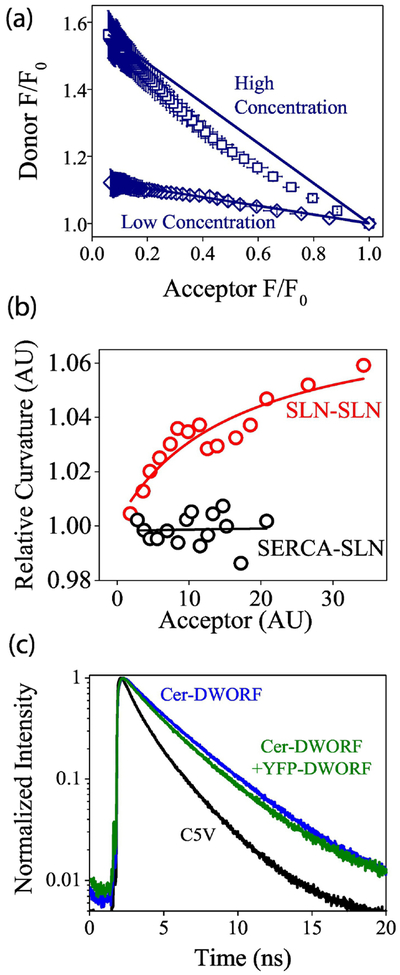
The micropeptide PLB forms avid pentamers, but previous studies have suggested that it is the monomeric form of PLB that binds and regulates the SERCA Ca transporter17. Similarly, SLN forms high-order (n>2) homo-oligomers but binds to SERCA as a monomer49. To determine whether the recently-discovered micropeptides formed regulatory heterodimers (or higher-order complexes) with SERCA, we performed acceptor-selective progressive acceptor photobleaching, as previously described17. Photobleaching of the YFP acceptor abolished energy transfer, dequenching the Cer donor and increasing its brightness (Fig. 3A). With progressive photobleaching of the acceptor (Fig. 3B, starting at arrow), we quantified the concomitant increase in donor fluorescence. Replotting progressive photobleaching data revealed the relationship between donor and acceptor fluorescence. A control plasmid with a Cerulean-Venus fusion construct at a fixed 1:1 donor:acceptor ratio (Fig. 3C, C5V)48 showed a linear donor vs. acceptor relationship, since every photobleached acceptor directly corresponds to one dequenched donor. In contrast, a control construct with two acceptors for each donor showed a highly curved donor-acceptor relationship (Fig. 3C, VCV). For this construct, FRET persists even after one of the acceptors is bleached, so donor dequenching lags behind acceptor photobleaching. A non-FRET control showed no donor dequenching after acceptor photobleaching (Fig. 3C, Cer-PLB+YFP). Applying progressive photobleaching analysis to the Cer-SERCA complex with YFP-micropeptides revealed linear donor-acceptor relationships for all micropeptides tested (Fig. 3D), indicating that, like PLB and SLN, the newly discovered micropeptides bind to SERCA as monomers.
Fig. 3.
Quantification of the interaction of SERCA2a with micropeptides using acceptor photobleaching. A) Acceptor (YFP-ALN) photobleaching increased donor (Cer-SERCA) fluorescence. B) Progressive acceptor photobleaching of the acceptor (starting at arrow) increased donor fluorescence. C) The relationship between donor and acceptor fluorescence reveals D:A stoichiometry. D) Progressive photobleaching suggested all micropeptides bind to SERCA as monomers.
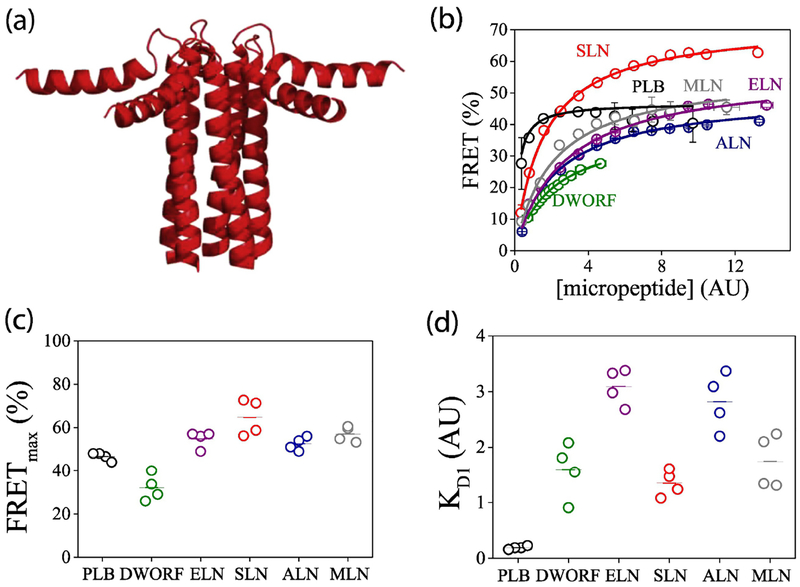
Micropeptides self-associate with different affinities
Since it is the monomeric form of the micropep-tides that interacts with SERCA, we considered it likely that possible oligomerization of the micropep-tides would be an important determinant of micro-peptide availability and functional potency. PLB is known to form pentamers (Fig. 4A) [50], and SLN is also proposed to form high-order oligomers [49] at he expense of the pool of monomers that bind SERCA. To quantify micropeptide oligomerization, we measured intra-oligomeric FRET by acceptor sensitization over a range of protein expression levels (Fig. 4B). Hyperbolic fits to the data revealed the FRETmax (Fig. 4C) and oligomer dissociation constant KD1 (Fig. 4D) for each micropeptide. We observed the lowest KD1 for PLB, with relative dissociation constants rank ordered as PLB < SLN = DWORF = MLN < ALN = ELN. We observed high cell–cell variability of FRET for MLN at all expression levels (Supplemental Fig. 3). We also noted that DWORF showed modest FRET in this assay (Fig. 4B), even though we did not detect coimmunoprecipitation of DWORF oligomers (Fig. 1B) and we previously did not observe significant DWORF-DWORF FRET [8]. This inconsistency may be due to the poor expression of this micropeptide (Figs. 2B, 4B, green), together with possible disruption of DWORF oligomers by detergent used in the present co-IP experiments. Other caveats relating to DWORF-DWORF interactions are discussed below.
Fig. 4.
Quantification of the homo-oligomerization of micropeptides by acceptor sensitization. A) The structure of the homopentamer of the micropeptide PLB. B) FRET increased with increasing protein expression, yielding estimates of homo-oligomer dissociation constants (KD1) and maximal FRET (FRETmax). C) Intrinsic FRET of micropeptide oligomer complexes. D) Apparent dissociation constants of micropeptide oligomers.
To evaluate the stoichiometry of micropeptide oligomers, we performed progressive acceptor photobleaching of 40–100 individual cells total, with 3–6 separate experiments per micropeptide (Supplementary Fig. 4). Interestingly, at low expression levels we observed a linear donor-acceptor relationship for all of the micropeptides. At increasing protein concentrations, the donor-acceptor plots became substantially non-linear. As an example, the donor-acceptor plots at high and low expression levels are shown for ALN in Fig. 5A, and results from other micropeptides are provided in Supplementary Fig. 5. After pooling data from cells of similar protein expression level, the degree of donor-acceptor relationship curvature was quantified from the area of the region bordered by the Cer/YFP data points and a line connecting the starting and ending points in the curve (Fig. 5A). The values obtained for SLN are plotted as a function of protein expression in Fig. 5B (red points), other micropeptide curvature analysis is provided in Supplementary Fig. 6. The plots revealed low (or no) curvature at low protein concentrations, increasing toward maximal curvature at high expression levels. This observation suggests that the micropeptide assembly pathway proceeds from monomers to dimers, then to higher order oligomers at high concentrations. For comparison, the SERCA-SLN complex showed photobleaching D:A plots with no curvature over a similar range of expression levels (Fig. 5B, black points).
Fig. 5.
Quantification of the homo-oligomerization of micropeptides by acceptor photobleaching and lifetime analysis. A) Progressive acceptor photobleaching showed that micropeptides (here ALN) formed dimers in low expressing cells, and higher order oligomers at high expression levels. B) Micropeptides (here SLN, red points) increasingly formed higher-order oligomers as expression increased, in contrast to the SERCA-SLN complex (black points). C) Fluorescence lifetime analysis of Cer-DWORF +YFP-DWORF fluorescence decay (green), compared to Cer-DWORF alone (blue) and a positive control high FRET construct, C5V (black).
DWORF, which showed weak expression and low FRET in acceptor sensitization experiments (Fig. 4C), also showed inconsistent results in acceptor photobleaching experiments. Specifically, some transfections resulted in some cells showing modest D:A photobleaching plot curvature (Supplementary Fig. 6), while other experiments showed nearly linear D:A photobleaching relationships. To determine the cause of the incongruent DWORF-DWORF FRET results, we performed time-correlated single photon counting analysis and observed a multiexponential decay for Cer-DWORF co-expressed with YFP-DWORF (Fig. 5C). The data are consistent with a majority (80%) non-FRET monomer population, with a smaller (20%) subpopulation of oligomers characterized by high FRET (62% FRET efficiency). This small fraction of oligomeric DWORF could account for the low overall FRET observed for this micropeptide.
Discussion:
Regulatory Complex Affinity and Stoichiometry-
The principal observation of the present study is that all of the micropeptides tested bind as monomers to SERCA. We do not see evidence of multiple acceptor-labeled micropeptides binding simultaneously to SERCA, as has been proposed for PLB42. However, we cannot rule out the possibility that oligomers bind SERCA in a configuration that puts all but one acceptor beyond reach of FRET. As we showed previously, both PLB and DWORF bind avidly to SERCA with comparable apparent affinities8, and here we determined that the other micropeptides have modestly lower affinity (higher KD2) for the pump. Overall, we regard these binding affinities as similar and compatible with the concept of mutual competition between micropeptides binding to SERCA in tissues where multiple species are expressed. Micropeptide-micropeptide FRET measurements suggest that all of the micropeptides can form oligomers, though with significantly lower self-affinity compared to PLB. The DWORF oligomerization data should be interpreted cautiously as the results were inconsistent, with evidence for dimers and higher order oligomers coexisting with a large population of monomers. MLN, SLN, and ALN FRET profiles were suggestive of positive cooperativity, and a Hill fit with Hill coefficients of >1 provided a better description of the data, particularly in the high protein concentration regime. Since we observed a linear D:A relationship during progressive photobleaching (Fig. 3D) we do not interpret this cooperativity as evidence of multiple binding sites for these micropeptides on SERCA. We have previously observed similar apparent positive cooperativity for some mutants of PLB23, which we attributed to depletion of the monomeric species by oligomerization at low protein expression levels. At high protein concentrations (>KD2), binding to SERCA increasingly competes with oligomerization, so FRET increases steeply. Our previous computational model predicted that such pseudocooperativity would be most noticeable when there is a large difference between the affinities of oligomerization and SERCA-binding43, but here we did not observe any particular relationship between the KD1/KD2 ratio and apparent positive cooperativity. One may also speculate that apparent cooperativity could result from micropeptides having different binding affinity for different conformations of SERCA (e.g. E1 vs. E2), as we have previously observed for phospholamban41. In live cells, different micropeptides may preferentially stabilize different enzymatic states of SERCA as it proceeds through the catalytic cycle, with apparently cooperative binding to the population overall.
SERCA-Micropeptide Regulatory Complex Quaternary Structure-
The relative SERCA-micropeptide donor-acceptor distances estimated here match what one might expect for the respective regulatory complexes. Specifically, the longer micropeptides like ALN and ELN showed higher FRETmax, likely because they can reach higher on the SERCA cytoplasmic headpiece, bringing the N-terminal YFP fusion closer to the Cer donor on the SERCA N-terminus in the actuator domain. Conversely, DWORF and SLN have smaller cytoplasmic domains, and lower FRETmax values. Compared to the other micropeptides, PLB and MLN showed unexpectedly low FRET for their size. In the case of PLB, this low FRET may be due to the propensity of the PLB cytoplasmic domain to interact dynamically with the surface of the phospholipid bilayer51–54. Such interactions are expected to decrease FRET from a donor fluorophore fused high on the opposite side of the SERCA cytoplasmic headpiece. Measured FRETmax values and calculated probe separation distances are provided in Table 1. For the purposes of this comparison, the distance calculations assume that the micropeptides bind to a single SERCA, however, our previous time-resolved spectroscopy experiments suggested that monomeric PLB may interact with a dimeric form of SERCA55. If the other micropeptides also bind to SERCA dimers in a similar conformation, the FRETmax measurements may overestimate the distance to the closest FRET partner.
Oligomer Architecture-
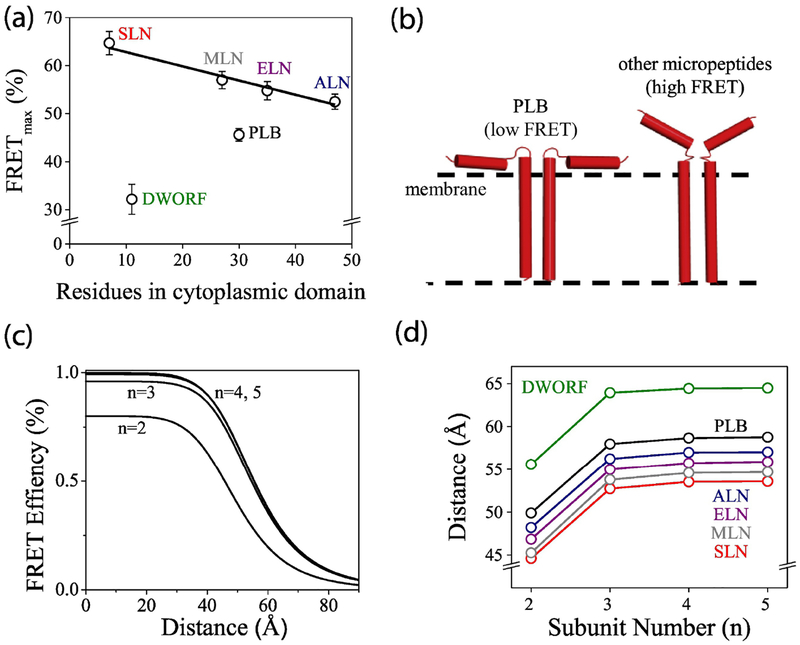
The structures of the micropeptide oligomers of the newly-discovered species have not been determined, but comparison of FRETmax values obtained for different micropeptides provides some insight into the overall quaternary conformation of the micropeptide homo-oligomers. We observed a general trend that larger micropeptides had lower intrinsic intra-oligomeric FRET (Fig. 6A). This is in harmony with the expectation that the longer the cytoplasmic domain, the greater the potential excursion of the N-terminal fluorescent protein tag from the central bundle of transmembrane helices, and therefore the larger the average separation distance between donors and acceptors. Notably, DWORF and PLB did not fall on this trendline. In particular, PLB had a lower intrinsic FRET than would be expected from the length of its cytoplasmic domain. We attribute this to the known propensity of PLB to interact with the surface of the membrane. While the structure of PLB is dynamic, fluorescence spectroscopy, NMR, and EPR demonstrate that the prevailing conformation of the PLB pentamer is one in which the cytoplasmic domains are splayed out onto the membrane surface50, 56–58 (Fig. 4A). This configuration creates maximal separation of the “arms” of the pentamer and reduces FRETmax. Such avid membrane interactions may be unique to PLB, and the cytoplasmic domains of the other micropeptides may not sustain such acute axial angles. Instead, they may spend relatively more time in close proximity as they sample the cytoplasmic space above the transmembrane helical bundle (Fig. 6B).
Fig. 6.
Interpretation of homo-oligomer FRET. A) For some micropeptides, oligomer intrinsic FRET decreased with increasing cytoplasmic domain size (black trend line). DWORF and PLB were outliers, suggesting unique conformations for these micropeptides. B) The data suggest that the cytoplasmic domains of PLB oligomers interact with the membrane surface more than those of other micropeptides. C) Theoretical FRET vs. distance relationships for ring-shaped oligomers of different stoichiometries. Trimers and tetramers cannot be distinguished from pentamers. D) The apparent probe separation distance calculated from FRETmax values of micropeptide homo-oligomers.
To interpret oligomer FRETmax values, we used a theory of FRET within a ring-shaped oligomer59 implemented as a Matlab model that incorporates the measured donor:acceptor ratio and assumes random assortment of donors and acceptors in the oligomer23. Fig. 6C shows that FRET depends strongly on probe separation distance and whether the oligomer is a dimer (n=2) or a higher order oligomer. High-order oligomer FRET curves (Fig. 6C, n=3, n=4,5) overlie one another because FRET cannot distinguish between trimers, tetramers, or pentamers for distances that are greater than the Förster distance R0 (49.8 Å for Cer-YFP). For those distances, the overall FRET is dominated by nearest-neighbor energy transfer, so distance estimates for oligomers >2 are indifferent to any number of additional protomers in the micropeptide oligomer (Fig. 6D).
Like PLB, DWORF showed an unexpectedly low FRETmax compared to the length of its predicted cytoplasmic domain (Fig. 6A). However, DWORF expressed poorly, and we were not able to explore the same range of protein concentrations as was examined for the other micropeptides (Fig. 4B). Thus, we have reduced confidence in the extrapolated maximum FRET value obtained from the hyperbolic fit of the binding curve. We also observed inconsistent results from the DWORF progressive photobleaching analysis (Supplementary Fig. 6B). Moreover, fluorescence lifetime analysis (Fig. 5C) suggested heterogeneity in the population of DWORF oligomer complexes. We suspect substantial populations of DWORF monomers and dimers persist even at higher protein expression levels. Significantly, if the DWORF FRETmax value is interpreted for a dimer FRET model, the DWORF-DWORF probe separation distance is similar to that of other micropeptides (Fig. 6D). Best estimates of the donor-acceptor separation distances, informed by likely oligomer stoichiometry, are highlighted in bold font in Table 1. DWORF is the only micropeptide species that does not inhibit SERCA (Fig. 1C), and it is noteworthy that it manifests unique oligomerization properties.
Interplay of SERCA-binding and Micropeptide Oligomerization-
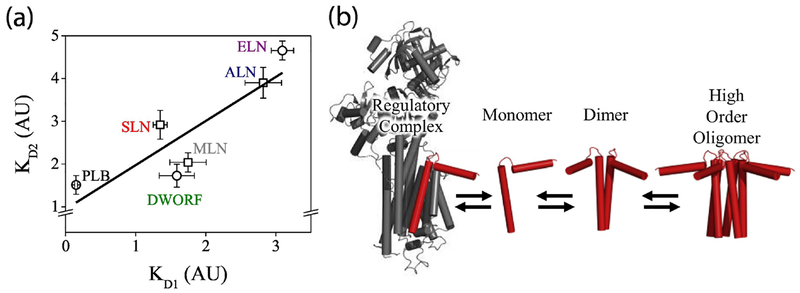
Based on previous studies that showed that depolymerization of PLB by mutations increased SERCA-binding16–19, we anticipated that there would be a negative correlation between oligomerization and SERCA-binding across the micropeptides investigated here. Surprisingly, comparison of each micropeptide’s oligomer dissociation constant (KD1) with that micropeptide’s apparent dissociation constant for SERCA (KD2) revealed a positive correlation (Fig. 7A). The fact that the most oligomeric species also bound SERCA well suggests that the same structural determinants that enable SERCA binding also confer oligomerization potential. Previously we have seen other examples of a positive KD2 vs. KD1 relationship, including a triple Ser substitution of PLB that worsened oligomerization and SERCA-binding28. Another previous PLB mutagenesis study showed a positive relationship between KD1 and KD2 for a series of ala substitutions, and a positive KD1/KD2 relationship for deletions of the C-terminal residues28. The data suggested that loss of structural elements that contribute to PLB oligomerization reduced the affinity of PLB for SERCA.
Fig. 7.
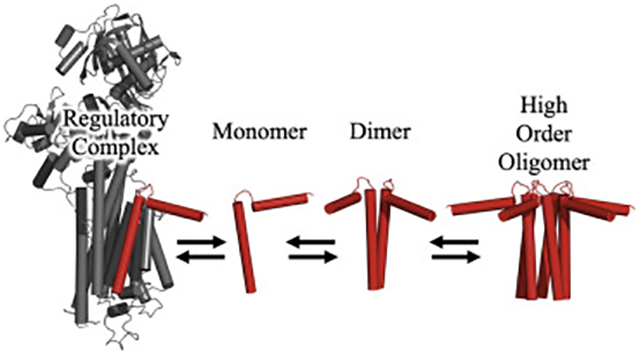
Summary of micropeptide regulatory interactions. A) SERCA-binding (KD2) correlated with oligomerization (KD1). Circles indicate KD2 obtained with a hyperbolic fit, squares indicate KD2 obtained with a Hill fit. B) A model of micropeptide interactions.
Micropeptide Oligomer Assembly Pathway-
Progressive photobleaching results suggest that all of the micropeptides are monomeric at low membrane concentrations. As the concentration increases, the first oligomeric species to form is a dimer, which proceeds to higher order oligomerization at increased micropeptide concentrations (Fig. 7B). This is not unexpected, as it is unlikely that a pentamer would spring from a simultaneous encounter of five appropriately oriented monomers. Stepwise assembly of oligomers is also suggested by previous SLN mutagenesis studies in which reducing SLN homo-oligomerization affinity also reduced oligomerization stoichiometry from a high-order species to a dimer49. Moreover, a pathogenic human mutation of PLB was shown to result in preformed PLB dimers due to oxidative cross-linking of an introduced cysteine (R9C). Such dimers had an enhanced ability to nucleate further oligomerization, such that WT-PLB increased its oligomerization in the presence of R9C-PLB24, 25.
Summary-
The present experiments shed new light on the diversity of micropeptides that regulate SERCA. Fig. 7B summarizes the conclusion that it is the monomer form of the inhibitory micropeptide (red) that interacts with the target ATPase (gray). This regulatory interaction occurs alongside stepwise oligomerization of micropeptides into dimers and higher order oligomers. The uniquely non-inhibitory micropeptide DWORF prevents binding of inhibitory micropeptides to SERCA, probably through competition for the inhibitory cleft in the SERCA transmembrane domain. Overall, the various micropeptides show similar biophysical properties, though PLB retains prime place. It is the most strongly oligomeric (Fig. 4D), it binds SERCA with the greatest affinity (Fig. 2D), and it shows the strongest functional impact on Ca transport (Fig. 1C). Future studies may reveal whether micropeptides that are differentially expressed in various tissues are appropriately optimized for interaction with the other SERCA isoforms. Overall, we envision a complex network of regulatory interactions of micropeptides, with specialized Ca handling in different cell types even within the same tissue. For example, in the heart there are cardiomyocytes that express PLB, SLN, ALN, and DWORF; endothelial cells that express ELN and ALN; and fibroblasts that express ALN-all expressing different SERCA isoforms. Moreover, it is unknown how the system is fine-tuned by post-translational modifications, or by micropeptide cross-talk interactions (e.g. hetero-oligomerization). Thus, we regard the present study of relative binding affinities a first step in understanding the rich complexity of micropeptide regulation of intracellular calcium handling.
Materials and Methods:
Plasmid Constructs
For all plasmid constructs, we used pEGFP-C1 as an expression vector in mammalian cells. All micropeptides (PLB, SLN, DWORF, ALN, ELN, and MLN) or SERCA constructs consisted of either mCerulean or EYFP fused via a 3 amino acid linker to the N-terminus of the micropeptide or SERCA8, 49, 60.
Cell culture and transfection
AAV 293 cells were cultured in DMEM cell culture medium supplemented with 10% fetal bovine serum (FBS) (ThermoScientific, Waltham, MA). Following culture, the cells were transiently transfected using MBS mammalian transfection kit (Agilent Technologies, Stratagene, La Jolla, CA) as per instructions provided with the kit. 24 hours post-transfection the cells were trypsinized (ThermoScientific, Waltham, MA) and replated onto poly-D-lysine coated glass bottom chambers and allowed to settle down for one hour before imaging.
Acceptor sensitization fluorescence resonance energy transfer
Imaging was performed using a wide-field fluorescent microscope as described previously41. Cells were imaged in 1 X PBS with magnesium (Hyclone Laboratories, Logan, Utah) using an inverted microscope (Nikon Eclipse Ti) equipped with a black-thinned EMCCD camera (iXon 887, Andor Technology, Belfast, Northern Ireland). The image acquisition of a field of view for each sample was performed with a 40 × 0.75 NA objective and 50 ms exposure time for Cer, YFP, and FRET (Cer excitation, YFP emission) channels. Metamorph software (Molecular Devices, Sunnyvale, CA) was used to quantify fluorescent intensity for the images collected in each channel. FRET efficiency was calculated according to E = G/(G + 3.2× FCer, where G=FFRET−a×FYFP−d×FCer61, where FFRET, FYFP, and FCer are the matching fluorescence intensity from FRET, YFP, and Cer images, respectively, and G represents FRET intensity corrected for the bleed-through of the channels. The parameters a and d are bleed-through constants calculated as a=FFRET/FCer for a control sample transfected with only YFP and d=FFRET/FYFP for a control sample transfected with only Cer. These values were determined to be G=4.74 a=0.075 and b=0.88. The FRET efficiency of each cell was plotted as a function of protein expression (measured from YFP fluorescence intensity). The relationship between FRET and expression was fit by a hyperbolic fit of the form FRET=(FRETmax)([micropeptide])/(Kd+[micropeptide]). For some micropeptides, FRET data did not fit well with a hyperbola, especially with respect to large values of X (high expressing cells), impeding estimation of FRETmax. For those micropeptides, data were fit with a Hill function of the form FRET=(FRETmax)([micropeptide]n)/(Kdn +[micropeptide]n) where n is the Hill coefficient. SERCA-micropeptide regulatory complex probe separation distance (r) was calculated from intrinsic FRET efficiency (FRETmax) according to the relationship described by T. Forster62: r = R0[(1/FRETmax – 1)1/6], where R0 is the Förster distance, which is 49.8 Å for the Cer-EYFP pair. The distance between fluorescent protein probes in micropeptide oligomers was calculated from FRETmax and the measured donor:acceptor ratio using a Matlab implementation of a model of FRET within a ring-shaped oligomer59 as previously described17, 23.
Progressive photobleaching
Progressive acceptor photobleaching was performed as described previously17. Briefly, we collected images of Cer and YFP fluorescence at intervals to establish a baseline and then initiated progressive acceptor photobleaching, acquiring successive images of Cer and YFP in between 10 sec of exposure to illumination through a 504/12 nm bandpass filter for selective photobleaching of YFP. The images were analyzed in Metamorph software (Molecular Devices, Sunnyvale, CA) and FRET was calculated from the pre- and post-bleach donor fluorescence intensity using the equation FRET=1-(FDA/FD), where FDA = the intensity of the donor before bleaching and FD = the intensity of the donor after bleaching. To distinguish between 1:1 and higher order stoichiometry, the fluorescence of the donor was plotted against the fluorescence of the acceptor at the same time point during progressive bleaching. A linear relationship was taken to indicate a dimer formation and supralinear relationship was taken to indicate a higher order oligomer formation of Cer- and YFP-labeled proteins17.
Time-correlated single photon counting (TCSPC)
TCSPC measurements were performed as previously described55. Pulsed excitation of Cer-DWORF was achieved with using a supercontinuum laser (Fianiam Ltd.) filtered through a heat mirror and a band-pass excitation filter of (427/10 nm), focused to a spot in the ER of HEK cells co-expressing Cer-DWORF and YFP-DWORF using a 60X water immersion objective. Fluorescence emission was collected through a band pass emission filter (472/30 nm) using an avalanche photodiode (Micro Photon Devices). Photon counting was performed with a PicoHarp 300 time-correlated single-photon counting module (PicoQuant Photonics, West Springfield, MA) and fluorescence decays were analyzed with SymPhoTime software. Cer-DWORF with no acceptor yielded a multiexponential decay63, so we considered several alternative approaches to interpretation of the decay data. Individual and global fits of donor alone and donor+acceptor samples with 2 or more exponentials showed that several of the components changed with addition of a FRET acceptor. This ambiguity prevented reliable measurement of apparent FRET distance. Qualitatively, the data suggested a small fraction of the donors were participating in FRET. To estimate the fraction and FRET efficiency of this subpopulation, we approximated the Cer lifetime as a single exponential fit, yielding a fluorescence lifetime of 3.45 ns. We then performed 2-exponential analysis of the FRET sample, fixing one lifetime at 3.45 ns, and observed a FRET subpopulation of 1.3 ns, compatible with 23.4 ± 1% of the donors having 62 ± 2% FRET efficiency (values are mean ± SE for n = 10 decays obtained from different cells).
Co-immunoprecipitation
CoIPs were performed as previously described3–5, 8. Briefly, HEK293 cells were co-transfected using FuGENE6 transfection reagent (Promega, Madison, Wisconsin) with expression plasmids encoding Myc-micropeptide and HA-micropeptide or Myc-SERCA2a and HA-micropeptide. Whole cell lysates were prepared in CoIP buffer (20 mM NaPO4, 150 mM NaCl, 2 mM MgCl2, 0.1% NP-40, 10% Glycerol, 10 mM sodium fluoride, 0.1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 1 mM DTT and Complete protease inhibitor (Roche, Basel, Switzerland). Immunoprecipitations were carried out using 1 mg of mouse monoclonal anti-Myc antibody (Invitrogen, Carlsbad, CA) and collected with Dynabeads (Invitrogen, Carlsbad, CA). Tris/Tricine gel electrophoresis was performed using pre-cast 16.5% Mini-PROTEAN TGX gels (Bio-Rad, Hercules, CA). Standard Western blot procedures were performed on input and IP fractions using the following antibodies: HA (Invitrogen, Carlsbad, CA), Myc (Invitrogen, Carlsbad, CA) or GAPDH (Millipore, Burlington, MA).
Oxalate-supported Ca2+ uptake
Oxalate-supported Ca2+ uptake in transfected HEK293 cells were measured as previously described in detail3–5, 8, 64. Cultured HEK293 cells were homogenized in 50 mM phosphate buffer, pH 7.0 containing 10 mM NaF, 1 mM EDTA, 0.3 M sucrose, 0.3 mM PMSF and 0.5 mM DTT. Ca2+ uptake was measured in reaction solution containing 40 mM imidazole pH 7.0, 95 mM KCl, 5 mM NaN3, 5 mM MgCl2, 0.5 mM EGTA, 5 mM K+ oxalate, 1 μM ruthenium red and various concentrations of CaCl2 to yield 0.02 to 5 μM free Ca2+. The reaction was initiated by the addition of ATP (final concentration 5 mM). The data were analyzed by nonlinear regression with computer software (GraphPad Software), and the KCa values were calculated using an equation for a general cooperative model for substrate activation.
Statistical Analysis
Data are presented as the mean ± standard error (S.E.) of n ≥ 3 experiments. All statical tests were performed using OriginPro 9.1 (OriginLab Corporation, Northampaton, MA). One-way analysis of variance (ANOVA) was used to compare the differences between groups. ANOVA was followed by Tukey’s post hoc test. A probability (p) value of < 0.05 was considered significant. Specific values are provided in figure panels or figure legends.
Supplementary Material
Highlights.
Micropeptides assemble stepwise as homo-dimers, then higher-order oligomers.
6 related micropeptides all bind to the SERCA2a calcium transporter as monomers.
Micropeptide oligomerization and SERCA-binding share structural determinants.
DWORF shows a unique profile of oligomerization/SERCA-binding.
Acknowledgements:
This work was supported by HL-092321, HL-143816, and support from the Loyola Stritch School of Medicine Cardiovascular Research Institute (to SLR); HL-130253, HD-087351, HL-138426, HL-092321, AR-067294, Fondation Leducq Networks of Excellence, and the Robert A Welch Foundation (grant 1–0025) (to ENO); NIH Pathway to Independence Award K99HL141630 (to CAM). The authors thank Sean R. Cleary, Aleksey V. Zima, Howard S. Young, and Gianluigi Veglia for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Tada M, Ohmori F, Yamada M, and Abe H (1979) Mechanism of the stimulation of Ca2+-dependent ATPase of cardiac sarcoplasmic reticulum by adenosine 3′:5′-monophosphate-dependent protein kinase. Role of the 22,000-dalton protein, J Biol Chem 254, 319–326. [PubMed] [Google Scholar]
- [2].Odermatt A, Taschner PE, Scherer SW, Beatty B, Khanna VK, Cornblath DR, Chaudhry V, Yee WC, Schrank B, Karpati G, Breuning MH, Knoers N, and MacLennan DH (1997) Characterization of the gene encoding human sarcolipin (SLN), a proteolipid associated with SERCA1: absence of structural mutations in five patients with Brody disease, Genomics 45, 541–553. [DOI] [PubMed] [Google Scholar]
- [3].Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, and Olson EN (2015) A micropeptide encoded by a putative long noncoding RNA regulates muscle performance, Cell 160, 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Anderson DM, Makarewich CA, Anderson KM, Shelton JM, Bezprozvannaya S, Bassel-Duby R, and Olson EN (2016) Widespread control of calcium signaling by a family of SERCA-inhibiting micropeptides, Sci Signal 9, ra119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, Reese AL, McAnally JR, Chen X, Kavalali ET, Cannon SC, Houser SR, Bassel-Duby R, and Olson EN (2016) A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle, Science 351, 271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Geering K, Beguin P, Garty H, Karlish S, Fuzesi M, Horisberger JD, and Crambert G (2003) FXYD proteins: new tissue- and isoform-specific regulators of Na,K-ATPase, Ann N Y Acad Sci 986, 388–394. [DOI] [PubMed] [Google Scholar]
- [7].Pirkmajer S, Kirchner H, Lundell LS, Zelenin PV, Zierath JR, Makarova KS, Wolf YI, and Chibalin AV (2017) Early vertebrate origin and diversification of small transmembrane regulators of cellular ion transport, J Physiol 595, 4611–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Makarewich CA, Munir AZ, Schiattarella GG, Bezprozvannaya S, Raguimova ON, Cho EE, Vidal AH, Robia SL, Bassel-Duby R, and Olson EN (2018) The DWORF micropeptide enhances contractility and prevents heart failure in a mouse model of dilated cardiomyopathy, Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ceholski DK, Trieber CA, and Young HS (2012) Hydrophobic imbalance in the cytoplasmic domain of phospholamban is a determinant for lethal dilated cardiomyopathy, J Biol Chem 287, 16521–16529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Negash S, Yao Q, Sun H, Li J, Bigelow DJ, and Squier TC (2000) Phospholamban remains associated with the Ca2+- and Mg2+-dependent ATPase following phosphorylation by cAMP-dependent protein kinase, The Biochemical journal 351, 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tada M, Inui M, Yamada M, Kadoma M, Kuzuya T, Abe H, and Kakiuchi S (1983) Effects of phospholamban phosphorylation catalyzed by adenosine 3′:5′-monophosphate- and calmodulin-dependent protein kinases on calcium transport ATPase of cardiac sarcoplasmic reticulum, J Mol Cell Cardiol 15, 335–346. [DOI] [PubMed] [Google Scholar]
- [12].Louis CF, Turnquist J, and Jarvis B (1987) Phospholamban stoichiometry in canine cardiac muscle sarcoplasmic reticulum, Neurochem Res 12, 937–941. [DOI] [PubMed] [Google Scholar]
- [13].Colyer J, and Wang JH (1991) Dependence of cardiac sarcoplasmic reticulum calcium pump activity on the phosphorylation status of phospholamban, J Biol Chem 266, 17486–17493. [PubMed] [Google Scholar]
- [14].Ferrington DA, Yao Q, Squier TC, and Bigelow DJ (2002) Comparable levels of Ca-ATPase inhibition by phospholamban in slow-twitch skeletal and cardiac sarcoplasmic reticulum, Biochemistry 41, 13289–13296. [DOI] [PubMed] [Google Scholar]
- [15].Kadambi VJ, Ponniah S, Harrer JM, Hoit BD, Dorn GW 2nd, Walsh RA, and Kranias EG (1996) Cardiac-specific overexpression of phospholamban alters calcium kinetics and resultant cardiomyocyte mechanics in transgenic mice, J Clin Invest 97, 533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Autry JM, and Jones LR (1997) Functional Co-expression of the canine cardiac Ca2+ pump and phospholamban in Spodoptera frugiperda (Sf21) cells reveals new insights on ATPase regulation, J Biol Chem 272, 15872–15880. [DOI] [PubMed] [Google Scholar]
- [17].Kelly EM, Hou Z, Bossuyt J, Bers DM, and Robia SL (2008) Phospholamban oligomerization, quaternary structure, and sarco(endo)plasmic reticulum calcium ATPase binding measured by fluorescence resonance energy transfer in living cells, J Biol Chem 283, 12202–12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kimura Y, Kurzydlowski K, Tada M, and MacLennan DH (1997) Phospholamban inhibitory function is activated by depolymerization, J Biol Chem 272, 15061–15064. [DOI] [PubMed] [Google Scholar]
- [19].Zvaritch E, Backx PH, Jirik F, Kimura Y, de Leon S, Schmidt AG, Hoit BD, Lester JW, Kranias EG, and MacLennan DH (2000) The transgenic expression of highly inhibitory monomeric forms of phospholamban in mouse heart impairs cardiac contractility, J Biol Chem 275, 14985–14991. [DOI] [PubMed] [Google Scholar]
- [20].Reddy LG, Jones LR, and Thomas DD (1999) Depolymerization of phospholamban in the presence of calcium pump: a fluorescence energy transfer study, Biochemistry 38, 3954–3962. [DOI] [PubMed] [Google Scholar]
- [21].Chen Z, Akin BL, and Jones LR (2007) Mechanism of reversal of phospholamban inhibition of the cardiac Ca2+-ATPase by protein kinase A and by anti-phospholamban monoclonal antibody 2D12, J Biol Chem 282, 20968–20976. [DOI] [PubMed] [Google Scholar]
- [22].Cornea RL, Jones LR, Autry JM, and Thomas DD (1997) Mutation and phosphorylation change the oligomeric structure of phospholamban in lipid bilayers, Biochemistry 36, 2960–2967. [DOI] [PubMed] [Google Scholar]
- [23].Hou Z, Kelly EM, and Robia SL (2008) Phosphomimetic mutations increase phospholamban oligomerization and alter the structure of its regulatory complex, J Biol Chem 283, 28996–29003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Abrol N, de Tombe PP, and Robia SL (2015) Acute inotropic and lusitropic effects of cardiomyopathic R9C mutation of phospholamban, J Biol Chem 290, 7130–7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ha KN, Masterson LR, Hou Z, Verardi R, Walsh N, Veglia G, and Robia SL (2011) Lethal Arg9Cys phospholamban mutation hinders Ca2+-ATPase regulation and phosphorylation by protein kinase A, Proc Natl Acad Sci U S A 108, 2735–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cornea RL, Autry JM, Chen Z, and Jones LR (2000) Reexamination of the role of the leucine/isoleucine zipper residues of phospholamban in inhibition of the Ca2+ pump of cardiac sarcoplasmic reticulum, J Biol Chem 275, 41487–41494. [DOI] [PubMed] [Google Scholar]
- [27].Zhai J, Schmidt AG, Hoit BD, Kimura Y, MacLennan DH, and Kranias EG (2000) Cardiac-specific overexpression of a superinhibitory pentameric phospholamban mutant enhances inhibition of cardiac function in vivo, J Biol Chem 275, 10538–10544. [DOI] [PubMed] [Google Scholar]
- [28].Abrol N, Smolin N, Armanious G, Ceholski DK, Trieber CA, Young HS, and Robia SL (2014) Phospholamban C-terminal residues are critical determinants of the structure and function of the calcium ATPase regulatory complex, J Biol Chem 289, 25855–25866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Periasamy M, Bhupathy P, and Babu GJ (2008) Regulation of sarcoplasmic reticulum Ca2+ ATPase pump expression and its relevance to cardiac muscle physiology and pathology, Cardiovasc Res 77, 265–273. [DOI] [PubMed] [Google Scholar]
- [30].Hasenfuss G, and Pieske B (2002) Calcium cycling in congestive heart failure, J Mol Cell Cardiol 34, 951–969. [DOI] [PubMed] [Google Scholar]
- [31].Medin M, Hermida-Prieto M, Monserrat L, Laredo R, Rodriguez-Rey JC, Fernandez X, and Castro-Beiras A (2007) Mutational screening of phospholamban gene in hypertrophic and idiopathic dilated cardiomyopathy and functional study of the PLN −42 C>G mutation, European journal of heart failure 9, 37–43. [DOI] [PubMed] [Google Scholar]
- [32].Haghighi K, Kolokathis F, Gramolini AO, Waggoner JR, Pater L, Lynch RA, Fan GC, Tsiapras D, Parekh RR, Dorn GW 2nd, MacLennan DH, Kremastinos DT, and Kranias EG (2006) A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy, Proc Natl Acad Sci U S A 103, 1388–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Schmitt JP, Kamisago M, Asahi M, Li GH, Ahmad F, Mende U, Kranias EG, MacLennan DH, Seidman JG, and Seidman CE (2003) Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban, Science 299, 1410–1413. [DOI] [PubMed] [Google Scholar]
- [34].Minamisawa S, Sato Y, Tatsuguchi Y, Fujino T, Imamura S, Uetsuka Y, Nakazawa M, and Matsuoka R (2003) Mutation of the phospholamban promoter associated with hypertrophic cardiomyopathy, Biochemical and biophysical research communications 304, 1–4. [DOI] [PubMed] [Google Scholar]
- [35].Kranias EG, and Hajjar RJ (2012) Modulation of cardiac contractility by the phospholamban/SERCA2a regulatome, Circulation research 110, 1646–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kawase Y, and Hajjar RJ (2008) The cardiac sarcoplasmic/endoplasmic reticulum calcium ATPase: a potent target for cardiovascular diseases, Nat Clin Pract Cardiovasc Med 5, 554–565. [DOI] [PubMed] [Google Scholar]
- [37].Hoshijima M (2005) Gene therapy targeted at calcium handling as an approach to the treatment of heart failure, Pharmacol Ther 105, 211–228. [DOI] [PubMed] [Google Scholar]
- [38].Greenberg B, Butler J, Felker GM, Ponikowski P, Voors AA, Desai AS, Barnard D, Bouchard A, Jaski B, Lyon AR, Pogoda JM, Rudy JJ, and Zsebo KM (2016) Calcium upregulation by percutaneous administration of gene therapy in patients with cardiac disease (CUPID 2): a randomised, multinational, double-blind, placebo-controlled, phase 2b trial, Lancet 387, 1178–1186. [DOI] [PubMed] [Google Scholar]
- [39].Asahi M, Sugita Y, Kurzydlowski K, De Leon S, Tada M, Toyoshima C, and MacLennan DH (2003) Sarcolipin regulates sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA) by binding to transmembrane helices alone or in association with phospholamban, Proc Natl Acad Sci U S A 100, 5040–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Akin BL, Hurley TD, Chen Z, and Jones LR (2013) The structural basis for phospholamban inhibition of the calcium pump in sarcoplasmic reticulum, J Biol Chem 288, 30181–30191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bidwell P, Blackwell DJ, Hou Z, Zima AV, and Robia SL (2011) Phospholamban binds with differential affinity to calcium pump conformers, J Biol Chem 286, 35044–35050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Glaves JP, Primeau JO, Espinoza-Fonseca LM, Lemieux MJ, and Young HS (2019) The Phospholamban Pentamer Alters Function of the Sarcoplasmic Reticulum Calcium Pump SERCA, Biophys J 116, 633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hou Z, and Robia SL (2010) Relative affinity of calcium pump isoforms for phospholamban quantified by fluorescence resonance energy transfer, J Mol Biol 402, 210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].King C, Stoneman M, Raicu V, and Hristova K (2016) Fully quantified spectral imaging reveals in vivo membrane protein interactions, Integr Biol (Camb) 8, 216–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sarabipour S, and Hristova K (2016) Mechanism of FGF receptor dimerization and activation, Nat Commun 7, 10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Singh DR, Ahmed F, Sarabipour S, and Hristova K (2017) Intracellular Domain Contacts Contribute to Ecadherin Constitutive Dimerization in the Plasma Membrane, J Mol Biol 429, 2231–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Singh DR, Kanvinde P, King C, Pasquale EB, and Hristova K (2018) The EphA2 receptor is activated through induction of distinct, ligand-dependent oligomeric structures, Commun Biol 1, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Koushik SV, Chen H, Thaler C, Puhl HL 3rd, and Vogel SS (2006) Cerulean, Venus, and VenusY67C FRET reference standards, Biophys J 91, L99–L101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Autry JM, Rubin JE, Pietrini SD, Winters DL, Robia SL, and Thomas DD (2011) Oligomeric interactions of sarcolipin and the Ca-ATPase, J Biol Chem 286, 31697–31706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Verardi R, Shi L, Traaseth NJ, Walsh N, and Veglia G (2011) Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method, Proc Natl Acad Sci U S A 108, 9101–9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Abu-Baker S, and Lorigan GA (2006) Phospholamban and its phosphorylated form interact differently with lipid bilayers: a 31P, 2H, and 13C solid-state NMR spectroscopic study, Biochemistry 45, 13312–13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Clayton JC, Hughes E, and Middleton DA (2005) The cytoplasmic domains of phospholamban and phospholemman associate with phospholipid membrane surfaces, Biochemistry 44, 17016–17026. [DOI] [PubMed] [Google Scholar]
- [53].Kirby TL, Karim CB, and Thomas DD (2004) Electron paramagnetic resonance reveals a large-scale conformational change in the cytoplasmic domain of phospholamban upon binding to the sarcoplasmic reticulum Ca-ATPase, Biochemistry 43, 5842–5852. [DOI] [PubMed] [Google Scholar]
- [54].Zamoon J, Mascioni A, Thomas DD, and Veglia G (2003) NMR solution structure and topological orientation of monomeric phospholamban in dodecylphosphocholine micelles, Biophys J 85, 2589–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Blackwell DJ, Zak TJ, and Robia SL (2016) Cardiac Calcium ATPase Dimerization Measured by Cross-Linking and Fluorescence Energy Transfer, Biophys J 111, 1192–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Pollesello P, Annila A, and Ovaska M (1999) Structure of the 1–36 amino-terminal fragment of human phospholamban by nuclear magnetic resonance and modeling of the phospholamban pentamer, Biophys J 76, 1784–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Robia SL, Flohr NC, and Thomas DD (2005) Phospholamban pentamer quaternary conformation determined by in-gel fluorescence anisotropy, Biochemistry 44, 4302–4311. [DOI] [PubMed] [Google Scholar]
- [58].Traaseth NJ, Verardi R, Torgersen KD, Karim CB, Thomas DD, and Veglia G (2007) Spectroscopic validation of the pentameric structure of phospholamban, Proc Natl Acad Sci U S A 104, 14676–14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Li M, Reddy LG, Bennett R, Silva ND Jr., Jones LR, and Thomas DD (1999) A fluorescence energy transfer method for analyzing protein oligomeric structure: application to phospholamban, Biophys J 76, 2587–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Robia SL, Campbell KS, Kelly EM, Hou Z, Winters DL, and Thomas DD (2007) Forster transfer recovery reveals that phospholamban exchanges slowly from pentamers but rapidly from the SERCA regulatory complex, Circulation research 101, 1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zal T, and Gascoigne NR (2004) Photobleaching-corrected FRET efficiency imaging of live cells, Biophys J 86, 3923–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Förster T (1948) Intermolecular energy migration and fluorescence, Ann. Phys. (Leipzig) 2, 55–75. [Google Scholar]
- [63].Sarkar P, Koushik SV, Vogel SS, Gryczynski I, and Gryczynski Z (2009) Photophysical properties of Cerulean and Venus fluorescent proteins, J Biomed Opt 14, 034047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Bidwell PA, and Kranias EG (2016) Calcium Uptake in Crude Tissue Preparation, Methods Mol Biol 1377, 161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.