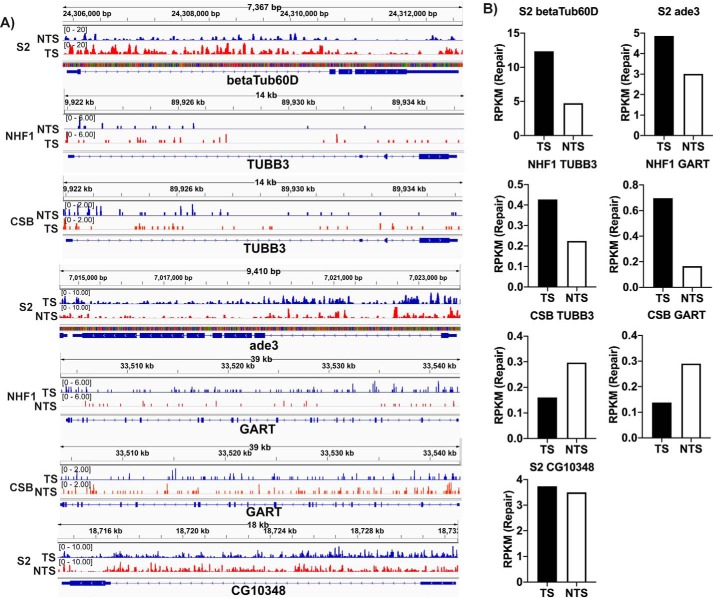
Figure 3.
Transcription-coupled repair in human NHF1 and Drosophila S2 cells at the single gene level. Screenshots (A) of XR-Seq data of select genes are shown along with quantitative analysis (B) of the read numbers. Genes analyzed include the betaTub60D gene from Drosophila and the human ortholog TUBB3, as well as the Drosophila ade3 gene and the human ortholog GART. Preferential repair in the TS as compared with the NTS of these genes is seen in the S2 and NHF1 cells but is absent in CSB cells, which in fact show the opposite preference, perhaps due to inhibition of TS repair by RNAP stalled at the damage. The Drosophila CG10348 gene is also analyzed in the bottom panels of A and B to illustrate an example of the absence of TCR. The y axis in each case is RPKM. The repair signal for Drosophila appears stronger because it has a much smaller genome and thus more reads per kbp per million reads. Time points are 30 min (S2 cells), 36 min (NHF1), and 1 h (CSB).