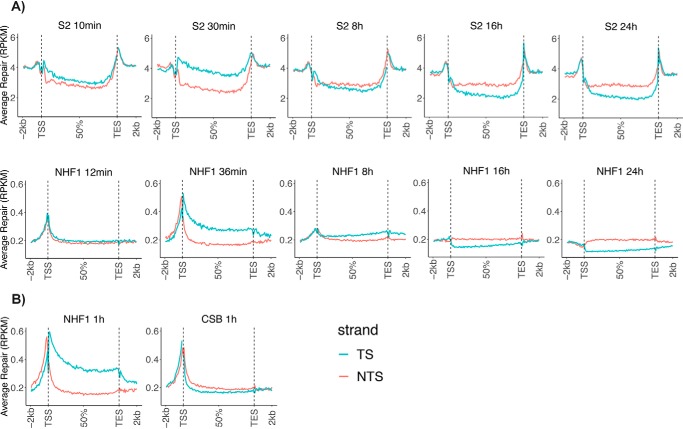
Figure 4.
Genome-wide analysis of TCR of transcribed genes in Drosophila and human cells. A, comparison of Drosophila S2 and human NHF1 cells. XR-Seq data from the indicated time points are plotted as average repair reads RPKM (y axis) along the length of a “unit gene.” For constructing a unit gene, a Drosophila data set of 5,706 genes was selected to include all genes >1 kbp with no genes that overlap or have a distance of less than 100 bp between adjacent genes. The human gene set includes 10,100 genes of >5 kbp in length with no overlaps and with a distance of at least 5 kbp between genes. For each species, the “unit gene” is 100 bins in length, and values for average repair were obtained by dividing each gene into 100 bins and averaging the repair values for each successive bin from 1 to 100. B, comparison of WT human NHF1 cells with cells from a patient with CSB. Data were analyzed as in A.