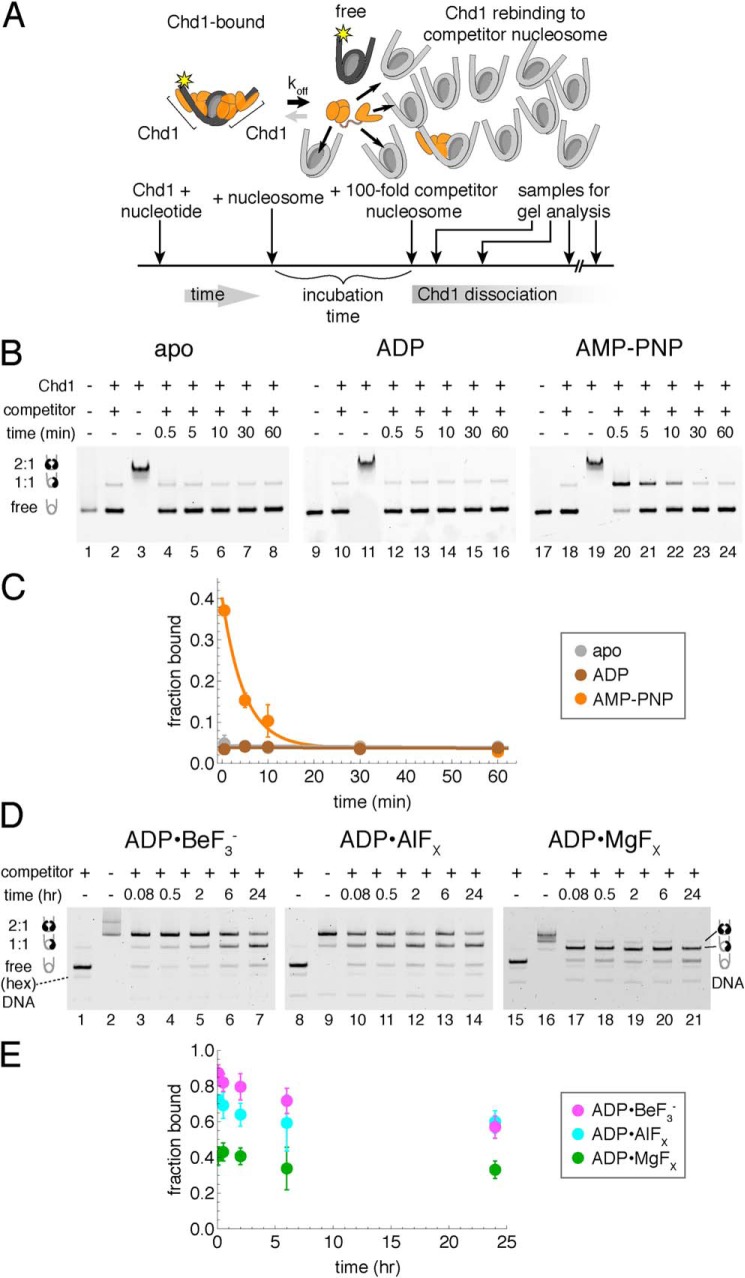
Figure 1.
A competition assay to determine stability of Chd1–nucleosome complexes. A, schematic workflow of a competition assay to determine off rates for Chd1. B, Chd1–nucleosome complexes are more stable in the presence of the nonhydrolyzable ATP analog AMP-PNP compared with ADP or no nucleotide (apo). As indicated, Chd1 (80 nm) was added to FAM-labeled 12N12 nucleosomes (20 nm), and after a 5-min incubation, reactions were competed with 12N9 unlabeled nucleosomes (2 μm). Lanes 2, 10, and 18 show reactions where unlabeled competitor nucleosome was first mixed with labeled nucleosome prior to addition of Chd1. Binding reactions were resolved on native acrylamide gels. Each gel is a representative of four or more experiments. C, quantification of Chd1–nucleosome dissociation over time. Disappearance of bound complexes in AMP-PNP was fit as a single exponential decay, giving an observed rate of 0.21 ± 0.03 min−1. Stably bound complexes were not detected under apo and ADP conditions. Each point represents the mean from four experiments, with error bars showing S.D. D, competition experiments reveal long-lived Chd1–nucleosome complexes in the presence of transition state analogs ADP·BeF3−, ADP·AlFX, and ADP·MgFX. Note that units of time are in hours. For these experiments, Chd1 (40 nm) was added to FAM-labeled 40N40 nucleosomes (10 nm), and after 2-h incubation, the reactions were competed with unlabeled 26N33 nucleosomes (1 μm). Lanes 1, 8, and 15 show reactions where unlabeled competitor nucleosome was first mixed with labeled nucleosome prior to addition of Chd1. Each gel is representative of five or more experiments. E, quantification of Chd1–nucleosome stability over time with transition state analogs. Each point represents an average of five or more experiments, with error bars showing S.D.