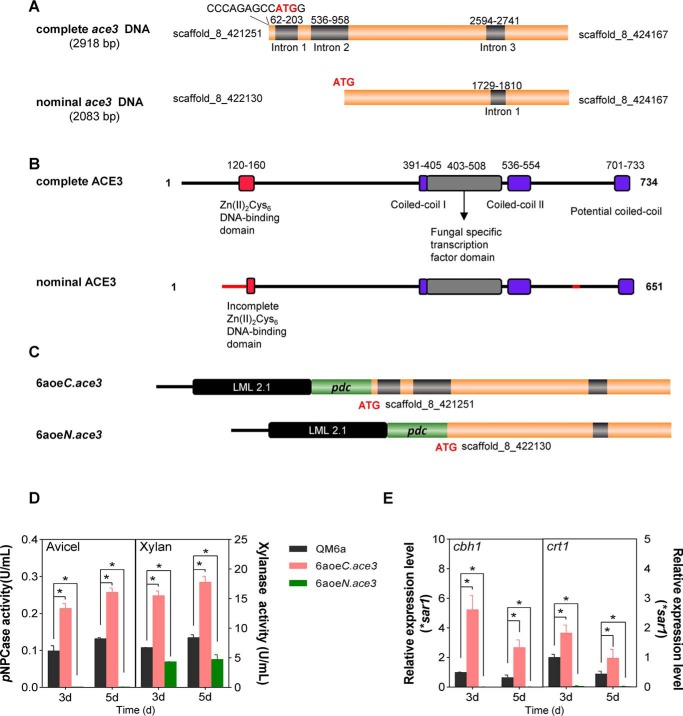
Figure 1.
Complete ACE3 structure and structural consequences of the nominal ACE3 sequence. A, complete and nominal ace3 sequences. B, the DNA-binding domain (red) of complete ACE3 is orthologous to that of Gal4 from Saccharomyces cerevisiae. Two coiled-coil (25) domains of ACE3 are indicated in purple. The potential coiled-coil domain of ACE3 is located at the C-terminal end of ACE3 (701–733 amino acids). The filamentous fungi-specific transcription factor domain (FSTFD; gray) regulates target gene expression. Hence, the nominal ACE3 sequence harbors the incomplete Zn(II)2Cys6 DNA-binding domain and erroneous N-terminal and C-terminal sequences (red line). The cDNA sequence of the complete ace3 sequence was submitted to GenBankTM (submission ID: BankIt2146245 ace3 MH819249). C, construction of two ace3 overexpression mutants. pdc is the strong constitutive promoter of pyruvate decarboxylase in T. reesei. LML 2.1 is the erasable hygromycin selection marker in T. reesei. Strain 6aoeC.ace3 is the complete ace3 overexpression mutant. Strain 6aoeN.ace3 is the nominal ace3 overexpression mutant. D, pNPCase and xylanase activities of T. reesei mutants 6aoeC.ace3 and 6aoeN.ace3 in Avicel and xylan, respectively. E, relative transcriptional levels of cbh1 and crt1 in T. reesei mutants in Avicel. The mRNA levels were normalized to the SAR1 housekeeping gene in each sample. Using the pdc promoter overexpressing the nominal ACE3 sequence totally blocked cellulase production compared with the ACE3 deletion strain (Fig. S2D). Overexpression of the complete ACE3 structure can increase the cellulase production (such as cbh1) and transcriptional levels of crt1 in T. reesei. Values are the mean ± S.D. (error bars) of the results from three independent experiments. Asterisks, significant difference compared with the untreated strain (*, p < 0.05, Student's t test).