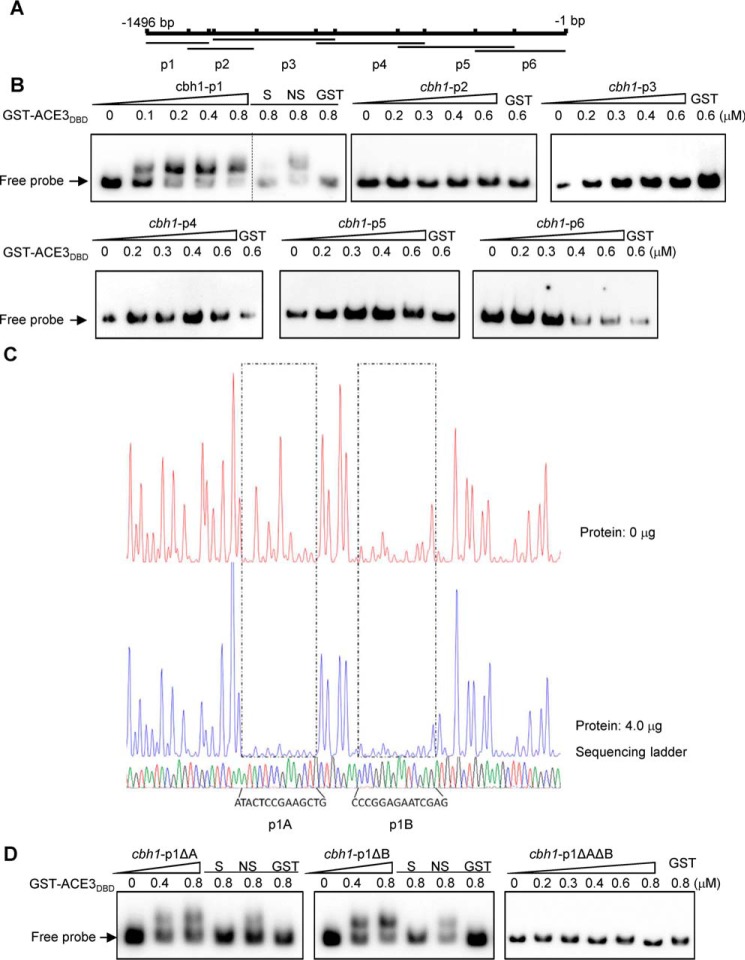
Figure 2.
ACE3 binds to cbh1 promoter in vivo and vitro. A, the cbh1 promoter was divided into the six parts, cbh1-p1, -p2, -p3, -p4, -p5, and -p6 fragments of the cbh1 promoter, and there was a 50–150-bp overlap between adjacent fragments, each of which were ∼200 to 450 bp long. B, the binding of GST-ACE3DBD (GST-ACE379–222) to the different cbh1 promoter fragments. EMSAs were performed by incubating biotin-labeled fragments cbh1-p1 (−1496 to −1272), cbh1-p2 (−1344 to −1102), cbh1-p3 (−1253 to −821), cbh1-p4 (−890 to −503), cbh1-p5 (−599 to −183), and cbh1-p6 (−322 to −1) of the cbh1 promoter with the GST tag–fused Zn(II)2Cys6 domain of ACE3 (aa 79–222) expressed by E. coli and purified by GST affinity chromatography. For each EMSA, a 10 nm biotin-labeled fragment and 0, 0.1, 0.2, 0.4, 0.6, and 0.8 μm (the increase amount is indicated by a right-pointing triangle) recombinant GST-ACE379–222 were added. EMSAs with only the GST tag (GST), a 200-fold excess of unlabeled specific fragments (S), or nonspecific competitor fragment (sperm DNA) (NS) were conducted as controls. C, electropherograms of the DNase I digest of the FAM-labeled cbh1-p1 fragment incubated without proteins (top of each panel) or with 4 μg of GST-ACE3DBD. The respective nucleotide sequences bound by GST-ACE3DBD are indicated as p1A and p1B (bottom of the panel). D, binding characteristics of GST-ACE3DBD to mutant cbh1-p1 fragments. EMSAs were identified by incubating biotin-labeled fragments cbh1-p1ΔA (deleting p1A), cbh1-p1ΔB (deleting p1B), and cbh1-p1ΔAΔB (deleting p1A and p1B). The dashed line indicates the splicing site of gels.