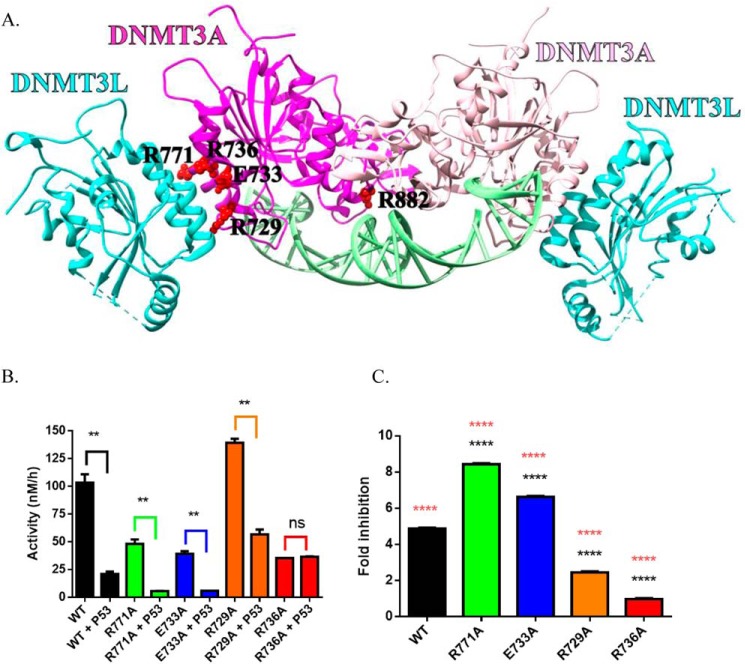
Figure 2.
DNMT3AWT tetramer interface mutants show highly variable response to p53WT inhibition. Crystal structure of a DNMT3AWT-DNMT3L complex (adapted from PDB code 5YX2) denoting critical residues for DNMT3AWT oligomerization (A) (36, 50). Although the extent of p53WT inhibition varies across DNMT3AWT mutants harboring a single alanine substitution within the tetramer interface, the DNMT3AR736A was unresponsive to p53WT inhibition (B and C). All reactions consisted of 150 nm DNMT3AWT and were initiated by the addition of 5 μm bp poly(dI-dC). For co-incubations, DNMT3AWT and p53WT (1:1) were preincubated for 1 h at 37 °C prior to starting the reaction by the addition of substrate DNA. Fold-inhibition was calculated by product formed by DNMT3A (WT and mutants) divided by product formed by DNMT3A (WT and mutants) without p53WT. All reactions were performed in duplicates. In B, a Student's unpaired t test was used to compare values within each set of reactions; **, p < 0.01; ns, p > 0.05. For C, a one-way analysis of variance was used to compare the values of each mutant to WT (****, p < 0.001) and across all samples (orange ****, p < 0.001). Data reflect the mean ± S.D. of 3 experiments.