Abstract
The PAH1-encoded phosphatidate phosphatase in Saccharomyces cerevisiae plays a major role in triacylglycerol synthesis and the control of phospholipid synthesis. For its catalytic function on the nuclear/endoplasmic reticulum membrane, Pah1 translocates to the membrane through its phosphorylation/dephosphorylation. Pah1 phosphorylation on multiple serine/threonine residues is complex and catalyzed by diverse protein kinases. In this work, we demonstrate that Pah1 is phosphorylated by the YCK1-encoded casein kinase I (CKI), regulating Pah1 catalytic activity and phosphorylation. Phosphoamino acid analysis coupled with phosphopeptide mapping of the CKI-phosphorylated Pah1 indicated that it is phosphorylated mainly on multiple serine residues. Using site-directed mutagenesis and phosphorylation analysis of Pah1, we identified eight serine residues (i.e. Ser-114, Ser-475, Ser-511, Ser-602, Ser-677, Ser-705, Ser-748, and Ser-774) as the target sites of CKI. Of these residues, Ser-475 and Ser-511 were specific for CKI, whereas the others were shared by casein kinase II (Ser-705), Cdc28–cyclin B (Ser-602), Pho85–Pho80 (Ser-114, Ser-602, and Ser-748), protein kinase A (Ser-667 and Ser-774), and protein kinase C (Ser-677). CKI-mediated phosphorylation of Pah1 stimulated both its phosphatidate phosphatase activity and its subsequent phosphorylation by casein kinase II. However, the CKI-mediated phosphorylation of Pah1 strongly inhibited its subsequent phosphorylation by Pho85–Pho80, protein kinase A, and protein kinase C. In a reciprocal analysis, Pah1 phosphorylation by Pho85–Pho80 inhibited subsequent phosphorylation by CKI. CKI-mediated Pah1 phosphorylation was also inhibited by a peptide containing the Pah1 residues 506–517, including the kinase-specific Ser-511 residue. These findings advance our understanding of how Pah1 catalytic activity and phosphorylation are regulated by multiple protein kinases.
Keywords: phospholipid, phosphatidic acid, diacylglycerol, triacylglycerol, phosphorylation, casein kinase I, PA phosphatase, Pah1, Yck1, yeast
Introduction
In the yeast Saccharomyces cerevisiae, the PAH1-encoded PAP4, 5 (1), which catalyzes the dephosphorylation of PA to yield DAG (2), has emerged as one of the most highly-regulated enzymes in lipid metabolism (3–7). Pah1 PAP activity largely governs whether cells utilize PA for the synthesis of membrane phospholipids via the liponucleotide CDP–DAG or for the synthesis of TAG via DAG (Fig. 1A) (3–6, 8). The impact of PAP activity on lipid metabolism extends beyond regulating the bifurcation of PA between CDP–DAG and DAG (8). By controlling the levels of PA and its derived lipids (1, 9, 10), the enzyme has a great influence on the expression of lipid synthesis genes (11, 12), nuclear/ER membrane growth (11), and lipid droplet formation (13). The PAP activity also impacts vacuole homeostasis (14), cell wall integrity (15, 16), TORC1-mediated induction of autophagy (17), growth on nonfermentable carbon sources (1, 18), susceptibility to fatty acid-induced lipotoxicity (9), sensitivity to heat (1, 11, 19, 20) and cold (21), and oxidative stress (22). Notably, cells lacking PAP activity have a shortened chronological life span (22) and exhibit apoptotic cell death in the stationary phase (9).
Figure 1.
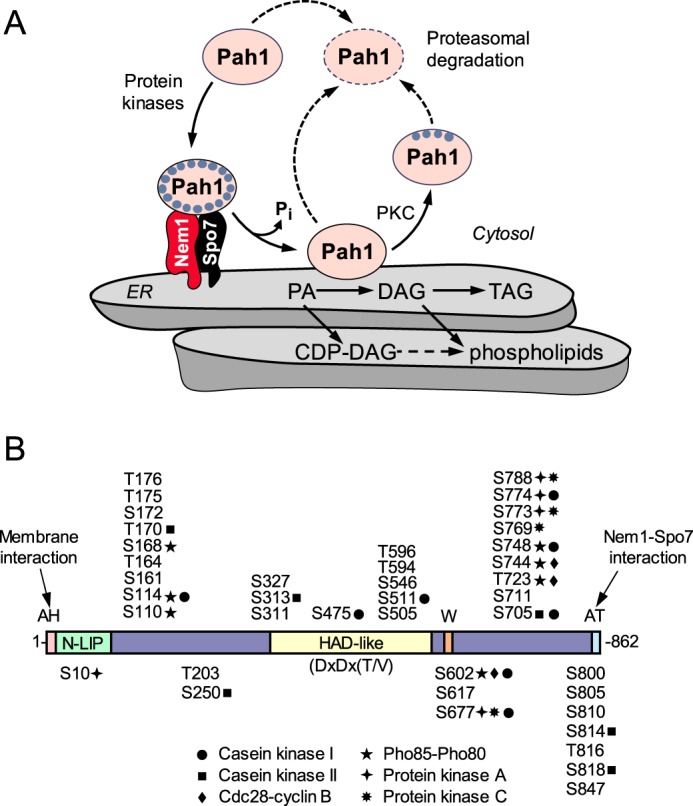
Model for the regulation of Pah1 PAP by phosphorylation, dephosphorylation, and proteasomal degradation, and domains/regions and phosphorylation sites in Pah1. A, Pah1 is phosphorylated by multiple protein kinases. The phosphorylated Pah1 (small blue circles) is recruited to the ER membrane through its association and dephosphorylation by the Nem1–Spo7 protein phosphatase complex (45, 49). Dephosphorylated Pah1 that is associated with the ER membrane via its amphipathic helix (45) catalyzes the conversion of PA to DAG (1), which is then acylated to form TAG. Dephosphorylated Pah1 or PKC-phosphorylated Pah1 that is not phosphorylated at the seven target sites for the Pho85–Pho80 protein kinase (42) is degraded by the 20S proteasome (indicated by the dashed line arrows) (48). PA is also utilized for the synthesis of membrane phospholipids via CDP–DAG (74, 107). When the CDP–DAG-dependent pathway for phospholipid synthesis is blocked, phosphatidylcholine or phosphatidylethanolamine may be synthesized from the DAG derived from the PAP reaction if cells are supplemented with choline or ethanolamine via the Kennedy pathway (74, 107). B, diagram shows the positions of the amphipathic helix (AH, pink) required for ER membrane interaction (45), the N-LIP (green) and HAD-like (yellow) domains that are required for PAP activity (18), the tryptophan (W) residue within the C-terminal conserved sequence WRDPLVDID (orange) required for function in vivo (108), and the acidic tail (AT, blue) required for interaction with the Nem1–Spo7 phosphatase complex (46). The serine (S) and threonine (T) residues known to be phosphorylated (38, 55–59, 59–65) are grouped at their approximate regions in the protein. The sites phosphorylated by CKI (this study), CKII (43), Cdc28–cyclin B (39), Pho85–Pho80 (40), PKA (41), and PKC (42) are indicated.
The PAP enzyme is conserved in eukaryotes, including humans (1, 23), mice (24, 25), flies (26, 27), worms (28), fungi (29), and plants (30, 31). All PAP enzymes have the haloacid dehalogenase (HAD)-like domain with a DXDX(T/V) catalytic motif and the N-LIP domain whose role in enzyme function is still unclear (Fig. 1B) (1, 18, 24). The critical roles that PAP plays in humans and mice are characterized by lipid-based syndromes associated with the loss or overexpression of the enzyme. For example, the deficiency of lipin 1 PAP in humans and mice causes rhabdomyolysis (32, 33), and its deficiency in mice is also characterized by hepatic steatosis during the neonatal period, lipodystrophy, insulin resistance, and peripheral neuropathy (24, 34). In contrast, the overexpression of lipin 1 PAP in mice causes an increase in lipogenesis and obesity (35). Moreover, the mutations of the human LPIN1 gene are associated with insulin resistance and the metabolic syndrome (36). Clearly, the regulation of the PAP expression and its catalytic activity is crucial for lipid homeostasis and cell physiology (5, 37).
Studies using yeast as a model organism have made great strides in understanding the mode of action and regulation of the PAP enzyme (5, 6, 8). Pah1 PAP is regulated by genetic and biochemical mechanisms, and its phosphorylation/dephosphorylation has emerged as a major biochemical regulation required to control its cellular function (3–6, 8). The posttranslational modification is essential for the subcellular localization of Pah1 and also affects its catalytic activity and protein stability (38–48). Pah1 phosphorylated by Pho85–Pho80 (40) and Cdc28–cyclin B (39) is less active due to its sequestration in the cytosol apart from the substrate PA in the nuclear/ER membrane (Fig. 1A). In addition, Pah1 PAP activity is diminished through its phosphorylation by the protein kinases at seven sites (Fig. 1B) distributed at the N and C termini of the protein (39, 40). In conjunction with Pho85–Pho80 and Cdc28–cyclin B, PKA functions to regulate the location and activity of Pah1 through its phosphorylation at five sites (Fig. 1B) distributed at the beginning and end of the protein (41). Moreover, Pah1 phosphorylated by those protein kinases is protected against its degradation by the 20S proteasome (47, 48). In contrast to its protection, Pah1 phosphorylated by PKC, which occurs on four sites at the C terminus (Fig. 1B), is susceptible to the proteasomal degradation (42). The phosphorylation of Pah1 by CKII, which occurs on six sites at the N and C termini, prevents its subsequent phosphorylation by PKA and stimulates its function in TAG synthesis (43). The regulatory effects of Pah1 phosphorylation by PKC (42) and CKII (43) on its stability and function are shown when it is not prephosphorylated at the seven sites by Pho85–Pho80/Cdc28–cyclin B. The phosphorylation of Pah1 by Pho85–Pho80/Cdc28–cyclin B exerts an inhibitory effect on its phosphorylation by PKC (42), but it has no effect on its phosphorylation by CKII (43). Unlike their effects on Pah1 phosphorylation, PKC (42) and CKII (43) have little effect on its PAP activity.
The phosphorylated form of Pah1, which is produced by multiple kinases (Fig. 1B), is dephosphorylated by a single protein phosphatase that is composed of the Nem1 (catalytic) and Spo7 (regulatory) subunits (44, 49). The catalytic activity of the membrane-bound Nem1–Spo7 phosphatase complex serves in translocating Pah1 to the nuclear/ER membrane; the dephosphorylated form of Pah1, which is associated with the membrane, is more active but prone to proteasomal degradation unlike its phosphorylated form produced by Pho85–Pho80, Cdc28–cyclin B, and PKA (38–41, 44, 45, 48). Based on our current understanding, Fig. 1A depicts a model for the phosphorylation/dephosphorylation-mediated regulation of Pah1 localization for its PAP function in lipid synthesis.
In this communication, we further examined the complex regulation of Pah1 by the CKI-mediated phosphorylation and found that Pah1 is a bona fide substrate of CKI, a serine/threonine protein kinase that plays roles in nutrient-mediated cell morphogenesis, cytokinesis, secretion, and endocytosis (50–54). Of eight serine residues of Pah1 phosphorylated by CK1, two residues (Ser-475 and Ser-511) were unique to the protein kinase and the others were common targets of phosphorylation by CKII, Pho85–Pho80, Cdc28–cyclin B, PKA, and PKC (Fig. 1B). The CKI phosphorylation of Pah1 stimulates its PAP activity and affects its subsequent phosphorylation by CKII, Pho85–Pho80, PKA, and PKC.
Results
Pah1 is a bona fide substrate of CKI
Pah1 is phosphorylated on at least 40 serine/threonine residues (Fig. 1B) (38, 55–65), but protein kinases that target the sites are not all known. The proteome-wide analysis of protein phosphorylation in vitro by Ptacek et al. (66) has indicated that Pah1 is also a target of protein kinases other than those already known (Fig. 1B). Among the protein kinases are Yck1 and Yck2, which are redundant yeast CKI isoforms (50, 51). Yck1 and Yck2, which associate with the plasma membrane via palmitoylation (67, 68), play redundant roles in cell physiology (50–53). In this work, we examined the phosphorylation of Pah1 by Yck1, which is more active on the substrate than Yck2 (66). To examine the CKI phosphorylation of Pah1, we utilized Yck1 purified from yeast and Pah1 purified from heterologous expression in Escherichia coli. The rationale for using the E. coli-expressed Pah1 was to examine its phosphorylation in the absence of endogenous phosphorylation in yeast (38). The affinity-purified Yck1 using a TAP tag was confirmed by immunoblot analysis with anti-protein A antibody (Fig. 2A). The phosphorylation of Pah1 by Yck1 was monitored by following the incorporation of the radioactive phosphate from [γ-32P]ATP into the substrate. In this analysis, the 32P-labeled Pah1 was resolved from [γ-32P]ATP by SDS-PAGE and then subjected to phosphorimaging. As shown in Fig. 2B (right), the E. coli-expressed Pah1 was phosphorylated by Yck1. Phosphoamino acid analysis of the 32P-labeled Pah1 showed that the protein is phosphorylated on serine (75%) and threonine (25%) residues with serine being a major target site (Fig. 2C). Phosphopeptide mapping of the 32P-labeled Pah1 indicates that the protein is phosphorylated by the CKI enzyme on multiple residues (Fig. 2D).
Figure 2.
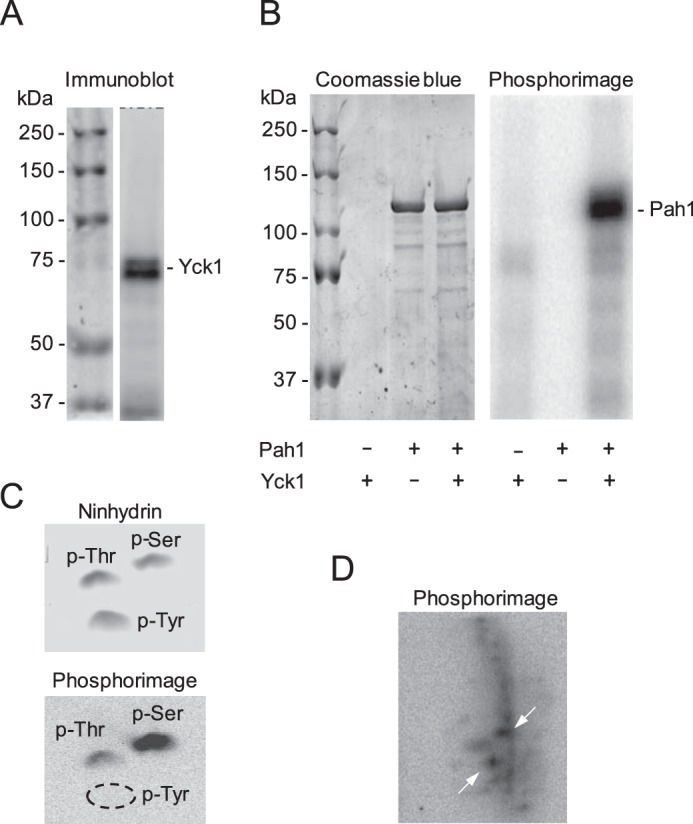
Pah1 is phosphorylated by CKI on multiple residues. A, the IgG–Sepharose-purified Yck1 preparation was subjected to immunoblot analysis using anti-protein A antibody. The positions of Yck1 and molecular mass standards are indicated. The doublet shown for Yck1 on the immunoblot may represent unphosphorylated and phosphorylated forms of the enzyme (59, 63, 65, 72). B, Pah1 (25 μg/ml) was incubated for 30 min at 30 °C in the presence (+) or absence (−) of 4 fmol/min Yck1 and 100 μm [γ-32P]ATP (3,000 cpm/pmol). The reaction mixtures were resolved by SDS-PAGE and subjected to phosphorimaging, followed by staining with Coomassie InstantBlue. The amount of Yck1 is too low for detection by protein staining of the polyacrylamide gel. The positions of Pah1 and the molecular mass standards are indicated. C, 32P-labeled Pah1 phosphorylated by CKI was incubated with 6 n HCl for 3 h at 100 °C. The acid hydrolysate was mixed with standard phosphoamino acids and resolved by two-dimensional electrophoresis on a cellulose TLC plate, which was subjected to phosphorimaging and staining with ninhydrin. The positions of phosphoserine (p-Ser), phosphothreonine (p-Thr), and phosphotyrosine (p-Tyr) as detected by ninhydrin staining are indicated. The dashed ellipse designates the absence of phosphotyrosine. D, 32P-labeled Pah1 phosphorylated by CKI was digested with TPCK-treated trypsin. The phosphopeptides produced by the proteolytic digestion were separated on cellulose TLC plates by electrophoresis (from left to right) in the first dimension and by chromatography (from bottom to top) in the second dimension; the TLC plate was subjected to phosphorimaging. The most heavily phosphorylated phosphopeptides are indicated by the white arrows. The data shown in all panels are representative of three experiments.
Using Pah1 as substrate, the CKI activity depended on reaction time and the amount of Yck1, indicating that the protein kinase follows zero-order kinetics (Fig. 3, A and B). Additionally, the CKI activity depended on the concentrations of Pah1 and ATP (Fig. 3, C and D). The Vmax and Km values for Pah1 are 16 fmol/min and 0.21 μm, respectively, and for ATP, the Vmax and Km values are 5.2 fmol/min and 2.4 μm, respectively. For both Pah1 (n = 1.8) and ATP (n = 2.7), the CKI activity followed positive cooperative kinetics, suggesting that the phosphorylation of one site enhances the phosphorylation of another site (69). Overall, these enzymological properties demonstrate that Pah1 is a bona fide substrate of Yck1 CKI.
Figure 3.
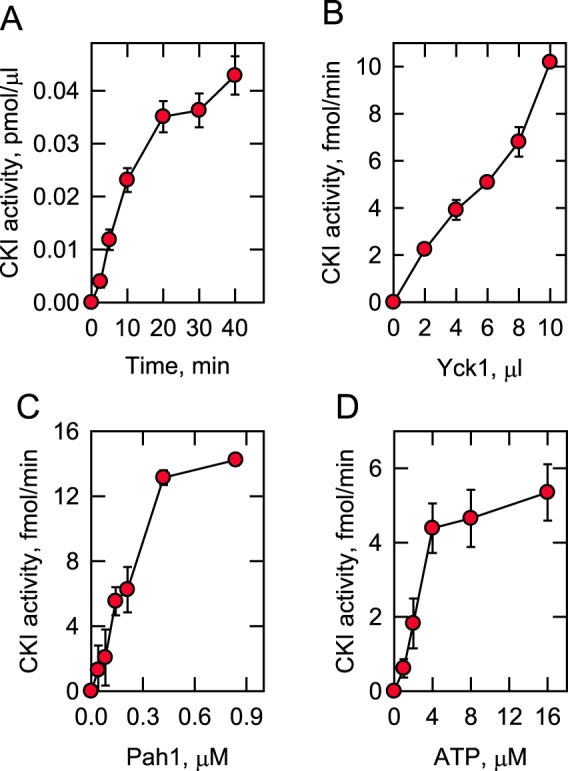
Phosphorylation of Pah1 by CKI depends on the reaction time and the amounts of CKI, Pah1, and ATP. Pah1 was incubated with Yck1 and [γ-32P]ATP at 30 °C. The phosphorylation of Pah1 was monitored by following the incorporation of the radioactive phosphate into the protein. After the reactions, the samples were subjected to SDS-PAGE to resolve the radioactive substrate and product, and the phosphorylated Pah1 was analyzed by phosphorimaging. The CKI phosphorylation reaction was conducted by varying the reaction time (A), the amount of Yck1 (B), and the concentrations of Pah1 (C) and ATP (D). A, B, and C, 100 μm ATP; B, C, and D, 30 min; A, B, and D, 25 μg/ml Pah1; A, C, and D, 5 fmol/min CKI. The data shown are means ± S.D. (error bars) from triplicate assays.
Mutational analysis of Pah1 identifies sites of phosphorylation by CKI
Sequence analysis of Pah1 using the NetPhosYeast (70) and NetPhos 3.1 (71) algorithms predicts multiple serine/threonine residues as target sites of phosphorylation by CKI. Of the putative sites of Pah1, 15 serine/threonine residues have been identified as being phosphorylated (Fig. 1B) (38, 41–43, 59, 72). Accordingly, we focused on these sites and examined the effects of their alanine substitutions on the phosphorylation of Pah1 (Fig. 4). Pah1 phosphorylation by CKI was significantly reduced by alanine substitutions for Ser-114, Ser-475, Ser-511, Ser-602, Ser-677, Ser-705, Ser-748, and Ser-774, indicating that the serine residues are the target sites of the protein kinase. The alanine substitution for Ser-511 showed the strongest reduction (90%), and those for Ser-475, Ser-748, and Ser-774 had the effect of >50% reduction in the CKI-mediated phosphorylation. Of those serine residues, Ser-475 (38) and Ser-511 (59, 72) are unique to CKI, whereas the others have also been identified as target sites for phosphorylation by CKII (Ser-705 (43)), Cdc28–cyclin B (Ser-602 (39)), Pho85–Pho80 (Ser-114, Ser-602, and Ser-748 (40)), PKA (Ser-677 and Ser-774 (41)), and PKC (Ser-677 (42)). Although Pah1 is also phosphorylated by CKI on the threonine residue (Fig. 2C), none of the threonine–to–alanine mutations examined (i.e. T93A, T124A, T153A, T157A, T170A, T176A, T221A, T234A, T315A, T353A, T364A, T517A, T553A, T662A, T723A, T778A, and T816A) had a significant effect on its phosphorylation. We reasoned that this might be because the extent of phosphorylation was much lower on the threonine residue than on the serine residue. Accordingly, we examined the mutational effect on the level of Pah1 phosphorylation on the threonine residue by phosphoamino acid analysis. However, none of the alanine substitutions for the threonine residues showed a significant reduction in Pah1 phosphorylation on the threonine residue.
Figure 4.
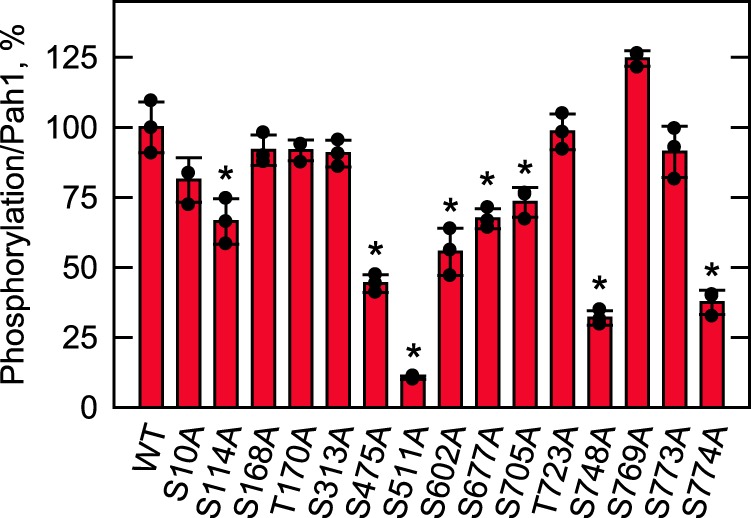
Phosphorylation site mutations reduce the phosphorylation of Pah1 by CKI. WT and the indicated Pah1 phosphorylation site mutants were expressed and purified from E. coli. The Pah1 mutant enzymes (25 μg/ml) were incubated for 30 min with 6 fmol/min CKI and 100 μm [γ-32P]ATP (3,000 cpm/pmol). Following the CKI phosphorylation reaction, Pah1 was separated from labeled ATP by SDS-PAGE and subjected to phosphorimaging and ImageQuant analyses. The amount of Pah1 used for each reaction was determined by ImageQuant analysis of the Coomassie Blue–stained SDS-polyacrylamide gel. The extent of phosphorylation of the WT enzyme was set at 100%. The data reported are the average of three independent experiments ± S.D. (error bars). The individual data points are also shown. *, p < 0.05 versus WT.
Pah1 phosphorylation by CKI increases its catalytic efficiency and relieves its cooperative dependence on PA
We questioned what effect the CKI phosphorylation of Pah1 has on its PAP activity. The CKI-mediated phosphorylation stimulated the PAP activity in a dose-dependent manner (Fig. 5A). Compared with unphosphorylated Pah1, the CKI-phosphorylated one exhibited ∼2-fold higher activity at its maximum stimulation. To gain insight into a mechanism for the stimulatory effect of the phosphorylation, we examined the unphosphorylated and phosphorylated forms of Pah1 as to their dependence of PAP activity on the surface concentration of PA in the Triton X-100/PA mixed micelle (1, 73). As expected (1), unphosphorylated Pah1 displayed positive cooperative kinetics (n = 2.7) with respect to PA (Fig. 5B). The phosphorylation by CKI relieved the cooperativity of the enzyme for its substrate; the phosphorylation reduced the Hill number for the reaction from 2.7 to 1.3. (Fig. 5B). Thus, the phosphorylated enzyme is more active at lower concentrations of PA when compared with the unphosphorylated enzyme. The Vmax values for the phosphorylated and unphosphorylated forms of Pah1 were 3.2 and 1.6 μmol/min/mg, respectively. The Km values for PA of the phosphorylated (Km = 4.1 mol %) and unphosphorylated (Km = 3.4 mol %) forms of Pah1 were not majorly different.
Figure 5.
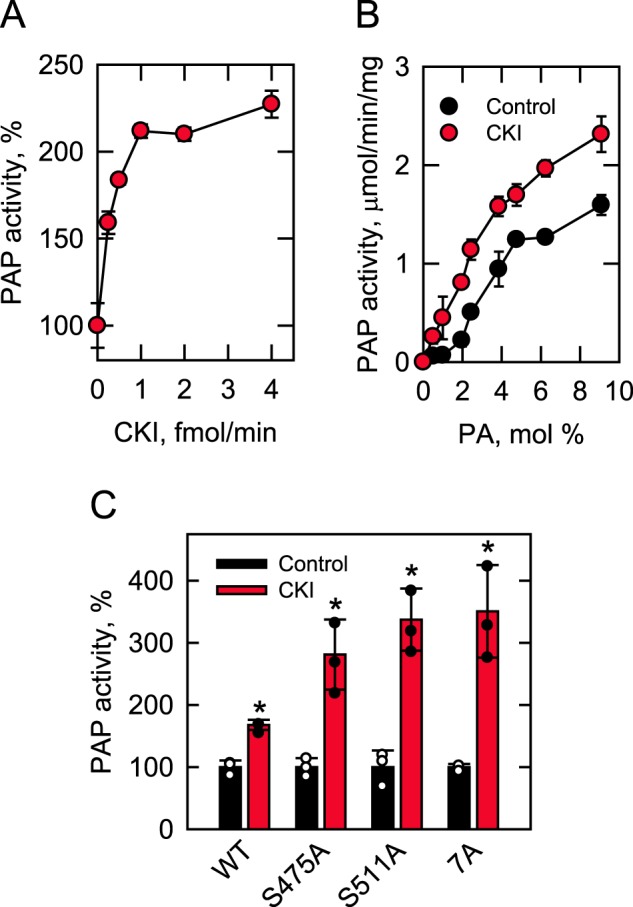
Phosphorylation of Pah1 by CKI stimulates PAP activity. Pah1 (10 μg/ml) was phosphorylated by CKI for 30 min with 100 μm ATP. After the phosphorylation, the PAP activity was measured by following the release of Pi from PA. The indicated amounts of CKI were used for the phosphorylation of Pah1 in A, whereas 4 fmol/min of CKI was used for the phosphorylation in B and C. PAP activity was measured with the indicated surface concentrations (mol %) of PA in B, whereas 9 and 2.4 mol % PA, respectively, were used for the measurement of PAP activity in A and C. The surface concentrations of PA were obtained by maintaining the molar concentration of PA at 0.2 mm and varying the molar concentration of Triton X-100 (73). The data shown are means ± S.D. (error bars) from triplicate assays. The individual data points are also shown in C. *, p < 0.05 versus control. Control, unphosphorylated; CKI, phosphorylated; S475A and S511A, CKI phosphorylation site mutations; 7A, Pho85–Pho80 phosphorylation site mutations.
The effect of the CKI-mediated phosphorylation on PAP activity was also examined with Pah1 mutant enzymes with the alanine substitutions for the unique CKI sites (Ser-475 and Ser-511) and the seven sites phosphorylated by Pho85–Pho80 (Fig. 5C). These experiments showed that the stimulation of PAP activity by CKI was not due to the phosphorylations at Ser-475, Ser-511, or the sites common to CKI and Pho85–Pho80. In fact, the phosphorylation of Pah1 with the S475A, S511A, and 7A mutations resulted in a greater stimulation (3–3.5-fold) of PAP activity when compared with the stimulation of activity caused by the phosphorylation of WT Pah1 (Fig. 5C).
Interrelationship of Pah1 phosphorylation between CKI and other protein kinases
We questioned whether the prephosphorylation of Pah1 by CKI affects its subsequent phosphorylation by CKII, Pho85–Pho80, PKA, and PKC. We did not examine the phosphorylation interrelationships with Cdc28–cyclin B because its three target sites are included in the seven sites phosphorylated by Pho85–Pho80 (Fig. 1B). Unphosphorylated Pah1, which had been prepared through heterologous expression in E. coli, was first phosphorylated by CKI with nonradioactive ATP and then phosphorylated by the other protein kinases with [γ-32P]ATP. Compared with a lack of prephosphorylation, the CKI-mediated prephosphorylation resulted in a dose-dependent stimulation of Pah1 phosphorylation by CKII (Fig. 6A). The prephosphorylation of Pah1 by 10 fmol/min CKI showed an 80-fold increase in its phosphorylation by CKII. The major sites of Pah1 phosphorylated by CKII include Thr-170, Ser-313, Ser-705, and Ser-818 (43). Pah-4A, which contains alanine substitutions for the four residues, did not exhibit the CKI-mediated stimulation of its phosphorylation by CKII (Fig. 5A), confirming that the increased phosphorylation was mediated by CKII. The alanine substitutions for Ser-475 and Ser-511, which are the CKI-specific target residues (Fig. 4), had major effects on phosphorylation by CKII (Fig. 6A). The S511A mutation reduced the CKI stimulatory effect by 80%, whereas the S475A mutation eliminated the stimulatory effect of prephosphorylation (Fig. 5A). These data support the conclusion that the CKI phosphorylation of Pah1 at Ser-475 and Ser-511 primes the subsequent phosphorylation by CKII. The CKI target sites of Pah1 that are also phosphorylated by Pho85–Pho80, PKA, and PKC are not expected to have a stimulatory effect on subsequent phosphorylation by CKII because the common site of prephosphorylation by the other kinases does not exhibit the stimulatory effect (43).
Figure 6.
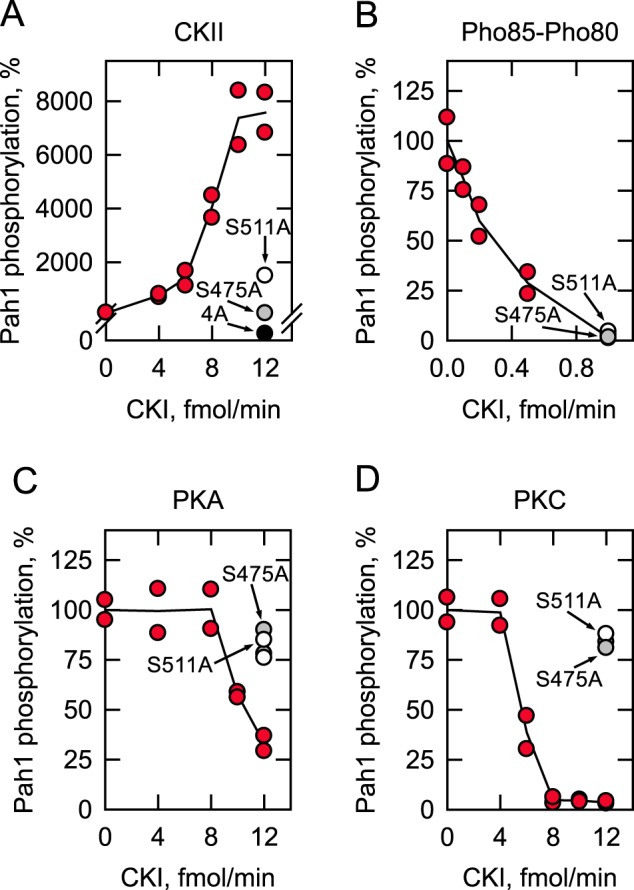
Prephosphorylation of Pah1 by CKI affects subsequent phosphorylations by CKII, Pho85–Pho80, PKA, and PKC. Unphosphorylated Pah1 (10 μg/ml) was prephosphorylated by the indicated amounts of CKI for 2 h with 100 μm ATP. The prephosphorylated Pah1 was then phosphorylated by 10 nmol/min CKII (A), 0.4 pmol/min Pho85–Pho80 (B), 4 pmol/min PKA (C), or 12 nmol/min PKC (D) with [γ-32P]ATP (3,000 cpm/pmol) for 2 h. The 32P-labeled Pah1 was separated from labeled ATP by SDS-PAGE and subjected to phosphorimaging and ImageQuant analysis. The amount of the phosphorylated Pah1 that was not subjected to prephosphorylation by CKI was set at 100%. The data reported are the results of two independent experiments, and the line drawn represents the average of the two experiments. S475A (gray circle) and S511A (white circle), CKI phosphorylation site mutations; 4A (black circle), and CKII phosphorylation site mutations.
In contrast to the stimulatory effect, Pah1 prephosphorylation by CKI showed a great inhibitory effect on its phosphorylations by Pho85–Pho80 (95%), PKA (67%), and PKC (95%) (Fig. 6, B–D). The inhibitory effect of prephosphorylation on the phosphorylation by Pho85–Pho80 could not be attributed to the CKI-specific phosphorylation sites (i.e. Ser-475 and Ser-511) because alanine substations for the serine residues did not alleviate the inhibitory effect (Fig. 6B). The inhibitory effect of the CKI-mediated Pah1 phosphorylation on the subsequent phosphorylation by Pho85–Pho80 might be explained by the fact that the two protein kinases share common phosphorylation sites (Fig. 1B). In addition, the amount of CKI needed to inhibit the subsequent phosphorylation by Pho85–Pho80 was 10-fold less than that needed to inhibit the subsequent phosphorylation by PKA and PKC or to stimulate the CKII-mediated phosphorylation. The CKI-mediated inhibition of Pah1 phosphorylation by PKA and PKC could be attributed to the phosphorylation at Ser-475 and Ser-511; the S475A and S511A mutations alleviated the inhibition caused by CKI (Fig. 6, C and D).
In reciprocal experiments, we examined whether the prephosphorylation of Pah1 by CKII, Pho85–Pho80, PKA, and PKC affects the subsequent phosphorylation by CKI. The prephosphorylation of Pah1 by Pho85–Pho80 caused a dose-dependent inhibition (75%) of its phosphorylation by CKI (Fig. 7B). However, the prephosphorylation of Pah1 by the other protein kinases had no or little effect on the subsequent phosphorylation of Pah1 by CKI (Fig. 7, A, C, and D). As discussed above, CKI and Pho85–Pho80 share common target sites (Fig. 1B), and thus the inhibitory effect of Pho85–Pho80 on the CKI activity was considered to be caused by a reduction in available target residues. To test this possibility, we analyzed the CKI phosphorylation of Pah1-7A, which has alanine substitutions for the seven target sites of Pho85–Pho80 (40). The 7A mutations reduced (40%) the inhibitory effect of the prephosphorylation by Pho85–Pho80 (Fig. 7B). Thus, the inhibition of the CKI-mediated phosphorylation is due to the phosphorylation by Pho85–Pho80 of sites common to both protein kinases. However, the data also indicate that phosphorylation by Pho85–Pho80 at sites that are not common to CKI affect the phosphorylation by CKI.
Figure 7.
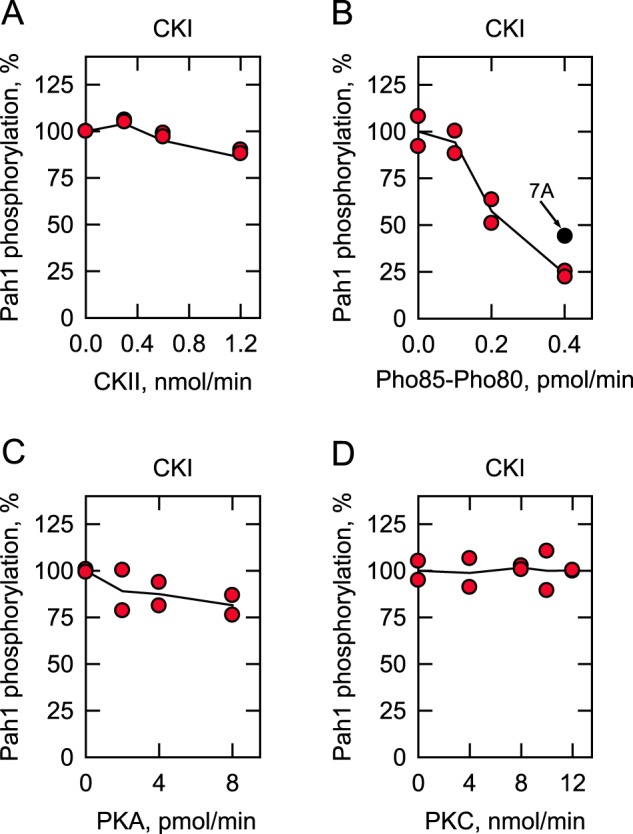
Effects of prephosphorylation of Pah1 by CKII, Pho85–Pho80, PKA, and PKC on subsequent phosphorylation by CKI. Unphosphorylated Pah1 (10 μg/ml) was prephosphorylated by the indicated amounts of CKII (A), Pho85–Pho80 (B), PKA (C), or PKC (D) for 2 h with 100 μm ATP. The prephosphorylated Pah1 was then phosphorylated by 6 fmol/min CKI with [γ-32P]ATP for 2 h. The 32P-labeled Pah1 was separated from labeled ATP by SDS-PAGE and subjected to phosphorimaging and ImageQuant analysis. The amount of the phosphorylated Pah1 that was not subjected to prephosphorylation by CKII, Pho85–Pho80, PKA, or PKC was set at 100%. The data reported are the result of two independent experiments, and the line drawn represents the average of the two experiments. 7A (black circle), and Pho85–Pho80 phosphorylation site mutations.
Pah1 phosphorylation by CKI is inhibited by a peptide containing Ser-511
Based on the highest reduction (90%) of Pah1 phosphorylation caused by the S511A mutation, Ser-511 was considered a major site of phosphorylation by CKI (Fig. 4). A peptide consisting of the Pah1 residues 506–517 was synthesized and examined as a substrate for CKI. In this analysis, the Pah1 peptide was phosphorylated by CKI with [γ-32P]ATP and captured on P81 phosphocellulose for radioactivity measurement by scintillation counting. The peptide served as a substrate of CKI, but the peptide phosphorylation was abolished by alanine substitution for Ser-511 (Fig. 8A). This result confirmed that Ser-511 of the peptide is the site of phosphorylation. Next, we questioned whether the peptide as a substrate competes with the full-length Pah1 for phosphorylation by CKI. The mixture of Pah1 and its peptide was phosphorylated by CKI with [γ-32P]ATP, and the phosphorylated Pah1 was resolved by SDS-PAGE and analyzed by phosphorimaging. The CKI phosphorylation of Pah1 was inhibited by its peptide (IC50 = 80 μm) in a dose-dependent manner (Fig. 8B). In contrast to the reduction of Pah1 phosphorylation, the peptide phosphorylation was reciprocally increased. These results indicate that the peptide is a competitive inhibitor of Pah1 phosphorylation by CKI (Fig. 8B). The Vmax and Km values for the Pah1 peptide were calculated to be 49 pmol/min/ml and 104 μm, respectively.
Figure 8.
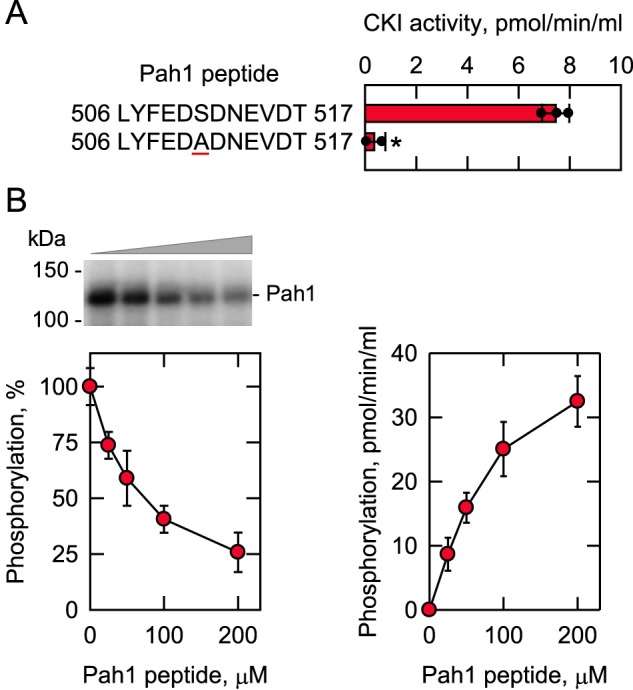
CKI phosphorylates the Pah1 synthetic peptide containing Ser-511; the synthetic peptide inhibits the phosphorylation of Pah1 by CKI. A, CKI activity was measured for 30 min with 100 μm of the indicated Pah1 synthetic peptides with 100 μm [γ-32P]ATP (5,000 cpm/pmol). The enzyme reaction was terminated by spotting the mixture onto P81 phosphocellulose paper, which was then washed with 75 mm phosphoric acid and subjected to scintillation counting. The numbers at the beginning and end of the peptides represent the positions in the full-length sequence of Pah1. The underlined residue within the peptide designates the serine–to–alanine substitution at position 511 in the sequence. The individual data points are also shown. B, Pah1 (1.25 μg/ml) was incubated for 30 min with 5 fmol/min CKI and 100 μm [γ-32P]ATP (5,000 cpm/pmol) in the absence and presence of the indicated concentrations of the WT Pah1 synthetic peptide. Following the CKI phosphorylation reaction, half of the sample was analyzed for the phosphorylation of Pah1 (left), and the other half was analyzed for the phosphorylation of the Pah1 peptide (right). Pah1 was separated from labeled ATP by SDS-PAGE and subjected to phosphorimaging and ImageQuant analysis. The extent of phosphorylation of the WT enzyme was set at 100%. The positions of Pah1 and the molecular mass standards are indicated. The phosphorylation of the Pah1 peptide was analyzed as described above. The data are the averages of three experiments ± S.D. (error bars). *, p < 0.05 versus WT. The phosphorimage shown in B (left) is representative of three experiments.
Discussion
Regulation of PAP activity is important for the control of lipid synthesis in eukaryotic organisms, including yeast (5, 37). The enzyme, which may govern whether cells utilize PA for the synthesis of the neutral lipid TAG or membrane phospholipids, is regulated on a biochemical level by the posttranslational modification of phosphorylation/dephosphorylation (3–6, 8, 74). The regulation of Pah1 by phosphorylation is complex; 40 phosphorylation sites have been identified, but only half of the sites can be ascribed to a specific protein kinase (Fig. 1B). Some Pah1 phosphorylation sites are common to multiple protein kinases (Fig. 1B), indicating that different signaling networks overlap to control the PAP function. This study focused on Pah1 phosphorylation by CKI, a constitutively active serine/threonine protein kinase that plays a key role in nutrient-mediated cell morphogenesis, cytokinesis, secretion, and endocytosis (50–54). The work here indicates that CKI also plays a role in lipid synthesis through the phosphorylation of Pah1. The enzymological studies demonstrated that Pah1 is a bona fide substrate of CKI; the phosphorylation reaction depended on time and the amounts of CKI, ATP, and Pah1. The Km value for Pah1 was essentially the same as that of CKII, Cdc28–cyclin B, and Pho85–Pho80 but was lower (2–3.5-fold) that that of PKA and PKC (Table 1). The Km value for ATP of the CKI reaction was lower (1.5–2.4-fold) than that of the other protein kinases known to phosphorylate Pah1 (Table 1). The stoichiometry of the CKI-mediated phosphorylation was 2.8 mol of phosphate/mol of Pah1, a value lower than that for the phosphorylation by Pho85–Pho80, but greater than that for the phosphorylation by other protein kinases (Table 1).
Table 1.
Kinetic properties of protein kinases that phosphorylate Pah1
The CKI phosphorylation of Pah1 had two major effects. First, Pah1 phosphorylated by the protein kinase was catalytically more active than the unphosphorylated form. The enzyme stimulation is shown to occur by a mechanism that increases the Vmax but reduces the positive cooperativity of the enzyme for PA. CKI is the first protein kinase that exhibits a stimulatory effect on the catalytic function of Pah1; other protein kinases (e.g. Pho85–Pho80 and PKA) inhibit its PAP activity (40, 41). Second, the CKI phosphorylation of Pah1 stimulated its phosphorylation by CKII but inhibited its phosphorylation by Pho85–Pho80, PKA, and PKC. The interrelationships of the phosphorylations are summarized in Fig. 9. The phosphorylation analysis of mutant Pah1 enzymes containing alanine substitutions for putative target serine residues led to the identification of eight CKI target sites with the major sites (based on ≥50% reduction in phosphorylation) being Ser-475, Ser-511, Ser-602, Ser-748, and Ser-774 (Fig. 4). The threonine residue(s) that is phosphorylated by CKI was not identified.
Figure 9.
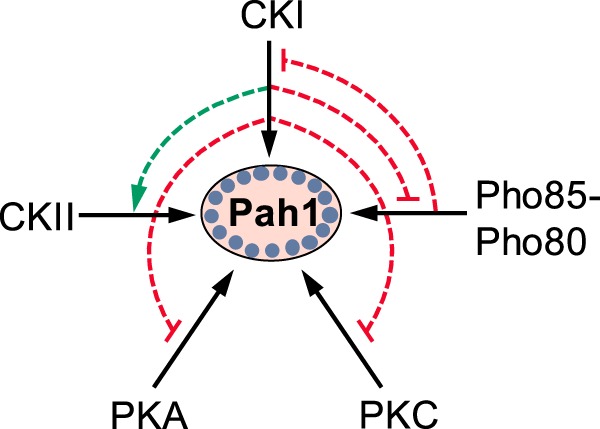
Interrelationships between the phosphorylation of Pah1 by CKI and phosphorylations by other protein kinases. Pah1 is phosphorylated by CKI, CKII, Pho85–Pho80, PKA, and PKC (solid black arrow). The CKI phosphorylation of Pah1 stimulates its subsequent phosphorylation by CKII (dashed green arrow) but inhibits its subsequent phosphorylations by Pho85–Pho80, PKA, and PKC (dashed blunted red line). The Pho85–Pho80 phosphorylation of Pah1 inhibits its subsequent phosphorylation by CKI (dashed blunted red line). Phosphorylated Pah1 is indicated by the small blue circles.
Six of the CKI sites are phosphorylated by other protein kinases. Being a constitutively-active protein kinase, CKI has the potential to phosphorylate a site(s) when another protein kinase is not active. For example, Pho85–Pho80 and PKA are most active during logarithmic growth when phospholipid synthesis is favored over TAG synthesis (75). Thus, at any point during the life cycle, lipid synthesis might be regulated via the CKI-mediated phosphorylation of Pah1 at a site(s) that is phosphorylated by another protein kinase that may not be active. This notion, however, has yet to be established in vivo.
Two serine residues (Ser-475 and Ser-511) of Pah1, which are located within the HAD-like domain (Fig. 1B), were identified as CKI-specific phosphorylation sites. The phosphorylation of these sites was shown to play roles in the subsequent phosphorylations by other protein kinases. On the one hand, the CKI phosphorylation of Ser-475 and Ser-511 was required for the stimulation of Pah1 phosphorylation by CKII. Previous work showed that the CKII phosphorylation of Pah1 stimulates the synthesis of TAG and lipid droplet formation, but only when it is not phosphorylated at the seven sites by Pho85–Pho80 (43). Thus, the CKI-mediated phosphorylation of Ser-475 and Ser-511 would be expected to have a positive impact on Pah1 function as mediated by CKII. On the other hand, the phosphorylation of Ser-475 and Ser-511 was required for the CKI-mediated inhibition of the phosphorylation by PKA and PKC. PKA works in conjunction with Pho85–Pho80 to attenuate Pah1 interaction with the ER membrane and inhibit PAP activity but at the same time to stabilize the enzyme to proteasomal degradation (41, 48). PKC facilitates the proteasomal degradation of Pah1, but only when the seven Pho85–Pho80 sites are not phosphorylated (42, 48). Accordingly, the phosphorylation of Ser-475 and Ser-511 might be expected to sustain the PKA- and PKC-mediated regulations of Pah1. The phosphorylation of Pah1 by CKI inhibited the subsequent phosphorylation by Pho85–Pho80, but the phosphorylation of Ser-475 and Ser-511 was not involved.
The phosphorylation of Ser-475 and Ser-511 was not involved in the stimulatory effect of CKI on PAP activity. In fact, the S475A and S511A mutations enhanced the stimulation of PAP by the CKI-mediated phosphorylation, suggesting that the phosphorylation of these sites inhibits the phosphorylation of another site(s) that must be responsible for the stimulation of activity. The phosphorylation of Pah1 by Pho85–Pho80 inhibits PAP activity, and the 7A mutation augmented the stimulatory effect CKI has on PAP activity. Clearly, the stimulation of the PAP activity by CKI is complex. The PAP activity stimulation observed with WT Pah1 is masked by inhibitory effects caused by the phosphorylation of some sites. Additional studies are needed to identify the phosphorylatable residue(s) responsible for the CKI-mediated stimulation of PAP activity.
The protein kinases (e.g. CKII, Pho85–Pho80, Cdc28–cyclin B, PKA, and PKC) previously known to phosphorylate Pah1 are associated with the soluble fraction (e.g. cytosol) of the cell (76). Accordingly, we presume that the phosphorylation of Pah1 by most protein kinases occurs in cytosol as depicted in Fig. 1A. However, the Yck1/2 isoforms of CKI associate with the plasma membrane through the posttranslational modification of palmitoylation (68), and their association with this membrane is required for their known physiological functions (77). Whether a soluble unmodified form of CKI phosphorylates Pah1 in the cytosol is unknown. Contact sites between the ER membrane and plasma membrane exist (78), and so CKI associated with the plasma membrane could phosphorylate Pah1 associated with the ER membrane. Additional studies are warranted to address this important question.
Like yeast Pah1, the mammalian lipin PAP enzymes are subject to regulation by phosphorylation at multiple serine/threonine residues (e.g. 44 sites in lipin 1β) (7, 79–84). Yet, only about 10% of the sites can be attributed to a specific protein kinase and signaling network (79, 80, 85, 86). It is known, however, that mouse lipin 1 is phosphorylated by mTORC1 (79, 80), CKI (86) and CKII (7). For example, stimulation by insulin is followed by the phosphorylation of lipin 1β by mTORC1 (79, 80, 85), whereas the phosphorylation by CKI causes lipin 1β to interact with the SCFβ-TRCP E3 ubiquitin ligase complex for its ubiquitination and degradation (86). In the case of lipin 1, the phosphorylation by CKI requires its prephosphorylation by mTORC1 (86). The phosphorylation of lipin 1 by CKII facilitates its interaction with 14-3-3 proteins for enzyme inactivation by retention in the cytoplasm apart from its substrate PA (7).
The lack of PAP activity in yeast, as well as in human, leads to aberrant lipid metabolism and cell physiology (5, 37). Too much PAP activity is also detrimental (e.g. obesity in the mouse model) (35). Thus, understanding the phosphorylation of PAP enzymes may lead to the identification of effector molecules to fine-tune activity and/or the enzyme cellular location. Peptide-based protein kinase inhibitors are attractive because they may block phosphorylation or disrupt protein–protein interactions (87, 88). Most peptide inhibitors of protein kinases are based on the consensus phosphorylation motif (87, 88). However, those inhibitors have a drawback of not having specificity for a particular substrate target of phosphorylation. A peptide sequence unique to a specific target like PAP would provide specificity without off-target side effects. The Pah1 peptide LYFEDSDNEVDT, which contains the major phosphorylation site Ser-511, inhibited the phosphorylation of Pah1 by CKI. This sequence is specific to Pah1; it does not align with any other proteins in the yeast database. The mechanism of inhibition was competitive with an IC50 value (80 μm) similar to the Km value (104 μm) for the peptide as a CKI substrate. Understandably, an effective inhibitor should work in the nanomolar range, and thus, the peptide tested in this work would not be useful. Yet, the data provide an initial proof–of–concept. Moreover, strategies (peptide cyclization and modification of backbone structure) are available that could enhance the inhibitory activity and effectiveness in vivo of the peptide (89). Studies with the Ser-511–containing peptide, as well as peptides that target other protein kinase target sites in Pah1, will be the subject of future work.
In summary, this work advances the understanding of the complex regulation of Pah1 PAP by phosphorylation. The CKI-mediated phosphorylation of Pah1 stimulates the PAP activity and regulates subsequent phosphorylations by CKII, Pho85–Pho80, Cdc28–cyclin B, PKA, and PKC. This study not only advances our knowledge of the multisite phosphorylations of Pah1, it also sheds new light on the CKI-mediated regulation of cellular processes.
Experimental procedures
Reagents
All chemicals used were reagent grade or better. Avanti Polar Lipids was the source of all lipids. Coomassie Blue R-250, DNA size ladders, molecular mass protein standards, and reagents for electrophoresis and immunoblotting were from Bio-Rad. Cayman Chemical was the source of leupeptin and pepstatin. Difco Laboratories was the supplier of all growth media. The InstantBlue protein stain was purchased from Expedeon. The Pah1 peptide was synthesized by EZBiolab. GE Healthcare was the source of IgG-Sepharose, Q-Sepharose, PVDF membrane, and the enhanced chemifluorescence Western blotting detection kit. MilliporeSigma was the source of ATP, l-1-tosylamido-2-phenylethyl chloromethyl ketone (TPCK)–trypsin, protease, and phosphatase inhibitors, phosphoamino acid standards, Triton X-100, Ponceau S stain, BSA, rabbit anti-protein A antibody (product no. P3775, lot no. 053M4806V), and cellulose and Silica Gel 60 TLC plates. Scintillation counting supplies and radiochemicals, respectively, were purchased from National Diagnostics and PerkinElmer Life Sciences. New England Biolabs was the source of enzyme reagents for DNA manipulations and human CKII. Promega was the source of bovine heart PKA catalytic subunit and rat brain PKC. Plasmid DNA purification kit and nickel-nitrilotriacetic acid–agarose resin were from Qiagen. The S. cerevisiae strain that expresses TAP-tagged Yck1 and the alkaline phosphatase-conjugated goat anti-rabbit IgG antibody (product no. 31340, lot no. NJ178812) were purchased from Thermo Fisher Scientific. Whatman supplied the P81 phosphocellulose paper.
Strains, plasmids, and growth conditions
The strains and plasmids used in this study are listed in Tables 2 and 3, respectively. E. coli strain DH5α was used for the propagation of plasmids. E. coli strains BL21(DE3)pLysS and BL21(DE3) were used for the induced expressions of Pah1 and Pho85–Pho80, respectively. Plasmid pGH313 directs the expression of His6-tagged Pah1 (1), and plasmids EB1164 and EB1076 direct the expressions of His6-tagged Pho85 and Pho80, respectively (90). The derivatives of pGH313 that contain threonine–to–alanine mutations were constructed by the QuikChange site-directed mutagenesis with appropriate templates and primers as described by Choi et al. (39). All mutations were confirmed by DNA sequencing. The isolation of plasmid DNA and its transformation into E. coli were performed by standard methods (91, 92). The bacterial cells were grown at 37 °C in lysogeny broth medium (1% tryptone, 0.5% yeast extract, 1% NaCl, pH 7.0). The growth medium was supplemented with ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) to select for cells carrying plasmids for the expression of proteins (93). The expression of proteins in E. coli cells was induced at 30 °C with 1 mm isopropyl β-d-1-thiogalactopyranoside. S. cerevisiae cells expressing TAP-tagged Yck1 were grown at 30 °C in YEPD medium (1% yeast extract, 2% peptone, and 2% glucose) (94). Cell numbers in liquid cultures were estimated by measuring absorbance at 600 nm.
Table 2.
Strains used in this study
| Strain | Genotype or relevant characteristics | Source or Refs. |
|---|---|---|
| E. coli | ||
| DH5α | F− φ80dlacZΔΜ15Δ (lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rk− mk+) phoA supE44 λ−thi-1 gyrA96 relA1 | 91 |
| BL21(DE3)pLysS | F− ompT hsdSB (rB−mB−) gal dcm (DE3) pLysS | Novagen |
| BL21(DE3) | F− ompT hsdSB (rB−mB−) gal dcm (DE3) | Invitrogen |
| S. cerevisiae | ||
| BY4741–YCK1–TAP | TAP-tagged Yck1 expressed in strain BY4741 | Thermo Fisher Scientific |
Table 3.
Plasmids used in this study
| Plasmid | Genotype or relevant characteristics | Source or Ref. |
|---|---|---|
| pET-15b | E. coli expression vector with N-terminal His6-tag fusion | Novagen |
| pGH313 | PAH1 coding sequence inserted into pET-15b | 1 |
| pGH313(S10A) | PAH1 (S10A) derivative of pGH313 | 41 |
| pGH313(T93A) | PAH1 (T93A) derivative of pGH313 | This study |
| pGH313(S114A) | PAH1 (S114A) derivative of pGH313 | 39 |
| pGH313(T124A) | PAH1 (T124A) derivative of pGH313 | This study |
| pGH313(T153A) | PAH1 (T153A) derivative of pGH313 | This study |
| pGH313(T157A) | PAH1 (T157A) derivative of pGH313 | This study |
| pGH313(S168A) | PAH1 (S168A) derivative of pGH313 | 39 |
| pGH313(T170A) | PAH1 (T170A) derivative of pGH313 | 43 |
| pGH313(T176A) | PAH1 (T176A) derivative of pGH313 | 43 |
| pGH313(T221A) | PAH1 (T221A) derivative of pGH313 | This study |
| pGH313(T234A) | PAH1 (T234A) derivative of pGH313 | This study |
| pGH313(S313A) | PAH1 (S313A) derivative of pGH313 | 43 |
| pGH313(T315A) | PAH1 (T315A) derivative of pGH313 | 43 |
| pGH313(T353A) | PAH1 (T353A) derivative of pGH313 | This study |
| pGH313(T364A) | PAH1 (T364A) derivative of pGH313 | 43 |
| pGH313(S475A) | PAH1 (S475A) derivative of pGH313 | 43 |
| pGH313(S511A) | PAH1 (S511A) derivative of pGH313 | 43 |
| pGH313(T517A) | PAH1 (T517A) derivative of pGH313 | 43 |
| pGH313(T553A) | PAH1 (T553A) derivative of pGH313 | 43 |
| pGH313(S602A) | PAH1 (S602A) derivative of pGH313 | 39 |
| pGH313(T662A) | PAH1 (T662A) derivative of pGH313 | This study |
| pGH313(S677A) | PAH1 (S677A) derivative of pGH313 | 41 |
| pGH313(S705A) | PAH1 (S705A) derivative of pGH313 | 43 |
| pGH313(T723A) | PAH1 (T723A) derivative of pGH313 | 39 |
| pGH313(S748A) | PAH1 (S748A) derivative of pGH313 | 39 |
| pGH313(S769A) | PAH1 (S769A) derivative of pGH313 | 42 |
| pGH313(S773A) | PAH1 (S773A) derivative of pGH313 | 41 |
| pGH313(S774A) | PAH1 (S774A) derivative of pGH313 | 41 |
| pGH313(T778A) | PAH1 (T778A) derivative of pGH313 | This study |
| pGH313(T816A) | PAH1 (T816A) derivative of pGH313 | This study |
| pGH313(4A) (CKII sites) | PAH1 (T170A/S313A/S705A/S818A) derivative of pGH313 | 43 |
| pGH313(7A) (Pho85-Pho80 sites) | PAH1 (S110A/S114A/S168A/S602A/T723A/S744A/S748A) derivative of pGH313 | 40 |
| EB1164 | PHO85-His6 derivative of pQE-60 | 90 |
| EB1076 | PHO80 derivative of pSBETA | 90 |
Enzyme preparations
All steps were performed at 4 °C. E. coli-expressed His6-tagged WT and mutant forms of yeast Pah1 (1) and His6-tagged Pho85–Pho80 protein kinase complex (90) were purified from bacterial cell extracts by affinity chromatography with nickel-nitrilotriacetic acid–agarose. The protein content of the enzyme preparations was estimated by the method of Bradford (95) using BSA as a standard. SDS-PAGE (96) analysis indicated that the Pah1 and Pho85–Pho80 preparations were highly purified. The TAP-tagged version of Yck1 was partially purified from the Triton X-100–solubilized extract (97) by affinity chromatography with IgG-Sepharose (98, 99). The cleavage with tobacco etch virus protease (98, 99) was omitted in the IgG-Sepharose chromatography step to retain the protein A tag on Yck1. Accordingly, the Yck1–TAP was eluted from the column with 50 mm glycine (pH 3.0) containing 0.1% Triton X-100 followed by the neutralization of the fractions by mixing with 0.2 volume of 1 m Tris-HCl (pH 8.0). The isolation of Yck1 was confirmed by immunoblotting with the anti-protein A antibody (Fig. 2A).
Protein kinase assays
Protein kinase activity was measured at 30 °C by following the incorporation of radiolabeled phosphate from [γ-32P]ATP into Pah1 in a total volume of 20 μl (39). The phosphorylation reactions were terminated by the addition of 6.7 μl of 4× Laemmli sample buffer (96). Phosphorylated Pah1 was resolved from labeled ATP by SDS-PAGE (96) and visualized by phosphorimaging. The extent of phosphorylation was quantified by ImageQuant software. For the phosphorylation of Pah1 peptide, the reaction was terminated by spotting the reaction mixture onto a P81 phosphocellulose paper, followed by phosphoric acid washing and scintillation counting. The reaction mixture for the protein kinase reactions contained 50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 100 μm [γ-32P]ATP (∼3,000 cpm/pmol), 40 μg/ml Pah1, 2 mm DTT, and the indicated amounts of the protein kinase. The reaction mixture for PKC also contained 1.7 mm CaCl2, 500 μm phosphatidylserine, and 156 μm DAG. The units of CKI, CKII, Pho85–Pho80, PKA, and PKC were defined as femtomoles/min, nanomoles/min, picomoles/min, picomoles/min, and nanomoles/min, respectively.
Phosphoamino acid analysis and phosphopeptide mapping
The 32P-labeled Pah1 transferred to PVDF membrane was hydrolyzed with 6 n HCl at 100 °C for phosphoamino acid analysis and digested with TPCK-trypsin for phosphopeptide mapping (100–102). The acid hydrolysates were mixed with standard phosphoamino acids and separated by two-dimensional electrophoresis on the cellulose plates. The tryptic digests were separated on the cellulose plates first by electrophoresis and then by TLC (100–102). Radioactive phosphoamino acids and phosphopeptides were visualized by phosphorimaging analysis. Nonradioactive phosphoamino acid standards were visualized by ninhydrin staining.
Immunoblotting
Proteins were separated by SDS-PAGE (96) using 8% slab gels. Immunoblotting with PVDF membrane was performed as described previously (103–105). Protein transfer from SDS-polyacrylamide gels to PVDF membranes was monitored by Ponceau S staining. Rabbit anti-protein A or rabbit anti-Pah1 (39) was used at a final concentration of 2 μg/ml. The secondary goat anti-rabbit IgG antibodies conjugated with alkaline phosphatase were used at a dilution of 1:5,000. Immune complexes were detected using the enhanced chemifluorescence immunoblotting substrate. Fluorimaging was used to acquire fluorescence signals from immunoblots, and the intensities of the images were analyzed by ImageQuant software. A standard curve was used to ensure that the immunoblot signals were in the linear range of detection.
PAP assay
PAP activity was measured by following the release of water-soluble Pi from chloroform-soluble PA using the Triton X-100/PA-mixed micellar assay as described by Han and Carman (23). The reaction mixture contained 50 mm Tris-HCl (pH 7.5), 1 mm MgCl2, 0.2 mm PA, 2 mm Triton X-100, and Pah1 protein in a total volume of 100 μl. Water-soluble Pi was measured with the malachite green–molybdate reagent at A650 nm (23, 106). Enzyme assays were conducted in triplicate, and the average standard deviation of the assays was ±5%. The enzyme reactions were linear with time and protein concentration. Protein concentration was estimated by the Coomassie Blue-based assay of Bradford (95) using BSA as standard. A unit of PAP activity was defined as micromoles/min.
Data analysis
Microsoft Excel software was used for the statistical analysis of data. The p values <0.05 were taken as a significant difference. The enzyme kinetics module of SigmaPlot software was used to analyze kinetic data according to Michaelis-Menten and Hill equations.
Author contributions
A. H., G.-S. H., and G. M. C. conceptualization; A. H. and G. M. C. data curation; A. H., G.-S. H., and G. M. C. formal analysis; A. H. and G. M. C. validation; A. H., L.-S. H., W.-M. S., G.-S. H., and G. M. C. investigation; A. H., L.-S. H., W.-M. S., G.-S. H., and G. M. C. visualization; A. H., L.-S. H., W.-M. S., G.-S. H., and G. M. C. methodology; A. H. writing-original draft; L.-S. H., W.-M. S., G.-S. H., and G. M. C. writing-review and editing; G. M. C. supervision; G. M. C. funding acquisition; G. M. C. project administration.
Acknowledgments
We thank Prabuddha Dey, Joanna Kwiatek, and Yeonhee Park for helpful discussions during this work.
This work was supported, in whole or in part, by National Institutes of Health Grant GM050679 from the USPHS. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PAP
- phosphatidate phosphatase
- CKI
- casein kinase I
- CKII
- casein kinase II
- CDP–DAG
- CDP-diacylglycerol
- DAG
- diacylglycerol
- PA
- phosphatidate
- PKA
- protein kinase A
- PKC
- protein kinase C
- PVDF
- polyvinylidene difluoride
- TAG
- triacylglycerol
- TLC
- thin-layer chromatography
- ER
- endoplasmic reticulum
- TPCK
- l-1-tosylamido-2-phenylethyl chloromethyl ketone
- HAD
- haloacid dehalogenase.
References
- 1. Han G.-S., Wu W.-I., and Carman G. M. (2006) The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281, 9210–9218 10.1074/jbc.M600425200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith S. W., Weiss S. B., and Kennedy E. P. (1957) The enzymatic dephosphorylation of phosphatidic acids. J. Biol. Chem. 228, 915–922 [PubMed] [Google Scholar]
- 3. Carman G. M., and Han G.-S. (2009) Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J. Biol. Chem. 284, 2593–2597 10.1074/jbc.R800059200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pascual F., and Carman G. M. (2013) Phosphatidate phosphatase, a key regulator of lipid homeostasis. Biochim. Biophys. Acta 1831, 514–522 10.1016/j.bbalip.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carman G. M., and Han G. S. (2019) Fat-regulating phosphatidic acid phosphatase: a review of its roles and regulation in lipid homeostasis. J. Lipid Res. 60, 2–6 10.1194/jlr.S087452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carman G. M. (2018) Discoveries of the phosphatidate phosphatase genes in yeast. J. Biol. Chem. 294, 1681–1689 10.1074/jbc.TM118.004159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hennessy M., Granade M. E., Hassaninasab A., Wang D., Kwiatek J. M., Han G.-S., Harris T. E., and Carman G. M. (2019) Casein kinase II-mediated phosphorylation of lipin 1β phosphatidate phosphatase at Ser-285 and Ser-287 regulates its interaction with 14-3-3β protein. J. Biol. Chem. 294, 2365–2374 10.1074/jbc.RA118.007246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kwiatek J. M., Han G. S., and Carman G. M. (2019) Phosphatidate-mediated regulation of lipid synthesis at the nuclear/endoplasmic reticulum membrane. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, S1388–1981(19)30042–3 10.1016/j.bbalip.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fakas S., Qiu Y., Dixon J. L., Han G.-S., Ruggles K. V., Garbarino J., Sturley S. L., and Carman G. M. (2011) Phosphatidate phosphatase activity plays a key role in protection against fatty acid-induced toxicity in yeast. J. Biol. Chem. 286, 29074–29085 10.1074/jbc.M111.258798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hassaninasab A., Han G.-S., and Carman G. M. (2017) Tips on the analysis of phosphatidic acid by the fluorometric coupled enzyme assay. Anal. Biochem. 526, 69–70 10.1016/j.ab.2017.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., and Siniossoglou S. (2005) The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24, 1931–1941 10.1038/sj.emboj.7600672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Han G.-S., and Carman G. M. (2017) Yeast PAH1-encoded phosphatidate phosphatase controls the expression of CHO1-encoded phosphatidylserine synthase for membrane phospholipid synthesis. J. Biol. Chem. 292, 13230–13242 10.1074/jbc.M117.801720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adeyo O., Horn P. J., Lee S., Binns D. D., Chandrahas A., Chapman K. D., and Goodman J. M. (2011) The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192, 1043–1055 10.1083/jcb.201010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sasser T., Qiu Q. S., Karunakaran S., Padolina M., Reyes A., Flood B., Smith S., Gonzales C., and Fratti R. A. (2012) The yeast lipin 1 orthologue Pah1p regulates vacuole homeostasis and membrane fusion. J. Biol. Chem. 287, 2221–2236 10.1074/jbc.M111.317420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lussier M., White A. M., Sheraton J., di Paola T., dwell J., Southard S. B., Horenstein C. I., Chen-Weiner J., Ram A. F., Kapteyn J. C., Roemer T. W., Vo D. H., Bondoc D. C., Hall J., Zhong W. W., et al. (1997) Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147, 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ruiz C., Cid V. J., Lussier M., Molina M., and Nombela C. (1999) A large-scale sonication assay for cell wall mutant analysis in yeast. Yeast 15, 1001–1008 [DOI] [PubMed] [Google Scholar]
- 17. Rahman M. A., Mostofa M. G., and Ushimaru T. (2018) The Nem1/Spo7-Pah1/lipin axis is required for autophagy induction after TORC1 inactivation. FEBS J. 285, 1840–1860 10.1111/febs.14448 [DOI] [PubMed] [Google Scholar]
- 18. Han G.-S., Siniossoglou S., and Carman G. M. (2007) The cellular functions of the yeast lipin homolog Pah1p are dependent on its phosphatidate phosphatase activity. J. Biol. Chem. 282, 37026–37035 10.1074/jbc.M705777200 [DOI] [PubMed] [Google Scholar]
- 19. Irie K., Takase M., Araki H., and Oshima Y. (1993) A gene, SMP2, involved in plasmid maintenance and respiration in Saccharomyces cerevisiae encodes a highly charged protein. Mol. Gen. Genet. 236, 283–288 10.1007/bf00277124 [DOI] [PubMed] [Google Scholar]
- 20. Han G.-S., O'Hara L., Carman G. M., and Siniossoglou S. (2008) An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J. Biol. Chem. 283, 20433–20442 10.1074/jbc.M802903200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Córcoles-Sáez I., Hernandez M. L., Martínez-Rivas J. M., Prieto J. A., and Randez-Gil F. (2016) Characterization of the S. cerevisiae inp51 mutant links phosphatidylinositol 4,5-bisphosphate levels with lipid content, membrane fluidity and cold growth. Biochim. Biophys. Acta 1861, 213–226 10.1016/j.bbalip.2015.12.014 [DOI] [PubMed] [Google Scholar]
- 22. Park Y., Han G. S., Mileykovskaya E., Garrett T. A., and Carman G. M. (2015) Altered lipid synthesis by lack of yeast Pah1 phosphatidate phosphatase reduces chronological life span. J. Biol. Chem. 290, 25382–25394 10.1074/jbc.M115.680314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han G.-S., and Carman G. M. (2010) Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J. Biol. Chem. 285, 14628–14638 10.1074/jbc.M110.117747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Péterfy M., Phan J., Xu P., and Reue K. (2001) Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27, 121–124 10.1038/83685 [DOI] [PubMed] [Google Scholar]
- 25. Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., and Reue K. (2007) Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 282, 3450–3457 10.1074/jbc.M610745200 [DOI] [PubMed] [Google Scholar]
- 26. Valente V., Maia R. M., Vianna M. C., and Paçó-Larson M. L. (2010) Drosophila melanogaster lipins are tissue-regulated and developmentally regulated and present specific subcellular distributions. FEBS J. 277, 4775–4788 10.1111/j.1742-4658.2010.07883.x [DOI] [PubMed] [Google Scholar]
- 27. Ugrankar R., Liu Y., Provaznik J., Schmitt S., and Lehmann M. (2011) Lipin is a central regulator of adipose tissue development and function in Drosophila. Mol. Cell. Biol. 31, 1646–1656 10.1128/MCB.01335-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Golden A., Liu J., and Cohen-Fix O. (2009) Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J. Cell Sci. 122, 1970–1978 10.1242/jcs.044743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu N., Yun Y., Yin Y., Hahn M., Ma Z., and Chen Y. (2019) Lipid droplet biogenesis regulated by the FgNem1/Spo7-FgPah1 phosphatase cascade plays critical roles in fungal development and virulence in Fusarium graminearum. New Phytol. 223, 412–429 10.1111/nph.15748 [DOI] [PubMed] [Google Scholar]
- 30. Nakamura Y., Koizumi R., Shui G., Shimojima M., Wenk M. R., Ito T., and Ohta H. (2009) Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc. Natl. Acad. Sci. U.S.A. 106, 20978–20983 10.1073/pnas.0907173106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eastmond P. J., Quettier A. L., Kroon J. T., Craddock C., Adams N., and Slabas A. R. (2010) Phosphatidic acid phosphohydrolase 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell 22, 2796–2811 10.1105/tpc.109.071423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zeharia A., Shaag A., Houtkooper R. H., Hindi T., de Lonlay P., Erez G., Hubert L., Saada A., de Keyzer Y., Eshel G., Vaz F. M., Pines O., and Elpeleg O. (2008) Mutations in LPIN1 cause recurrent acute myoglobinuria in childhood. Am. J. Hum. Genet. 83, 489–494 10.1016/j.ajhg.2008.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang P., Verity M. A., and Reue K. (2014) Lipin-1 regulates autophagy clearance and intersects with statin drug effects in skeletal muscle. Cell Metab. 20, 267–279 10.1016/j.cmet.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nadra K., de Preux Charles A.-S., Médard J.-J., Hendriks W. T., Han G.-S., Grés S., Carman G. M., Saulnier-Blache J.-S., Verheijen M. H., and Chrast R. (2008) Phosphatidic acid mediates demyelination in Lpin1 mutant mice. Genes Dev. 22, 1647–1661 10.1101/gad.1638008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Phan J., and Reue K. (2005) Lipin, a lipodystrophy and obesity gene. Cell Metab. 1, 73–83 10.1016/j.cmet.2004.12.002 [DOI] [PubMed] [Google Scholar]
- 36. Wiedmann S., Fischer M., Koehler M., Neureuther K., Riegger G., Doering A., Schunkert H., Hengstenberg C., and Baessler A. (2008) Genetic variants within the LPIN1 gene, encoding lipin, are influencing phenotypes of the metabolic syndrome in humans. Diabetes 57, 209–217 10.2337/db07-0083 [DOI] [PubMed] [Google Scholar]
- 37. Reue K., and Wang H. (2019) Mammalian lipin phosphatidic acid phosphatases in lipid synthesis and beyond: metabolic and inflammatory disorders. J. Lipid Res. 60, 728–733 10.1194/jlr.S091769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O'Hara L., Han G.-S., Peak-Chew S., Grimsey N., Carman G. M., and Siniossoglou S. (2006) Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem. 281, 34537–34548 10.1074/jbc.M606654200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Choi H.-S., Su W.-M., Morgan J. M., Han G.-S., Xu Z., Karanasios E., Siniossoglou S., and Carman G. M. (2011) Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of Ser602, Thr723, and Ser744 as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J. Biol. Chem. 286, 1486–1498 10.1074/jbc.M110.155598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choi H.-S., Su W.-M., Han G.-S., Plote D., Xu Z., and Carman G. M. (2012) Pho85p–Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J. Biol. Chem. 287, 11290–11301 10.1074/jbc.M112.346023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Su W.-M., Han G.-S., Casciano J., and Carman G. M. (2012) Protein kinase A-mediated phosphorylation of Pah1p phosphatidate phosphatase functions in conjunction with the Pho85p–Pho80p and Cdc28p–cyclin B kinases to regulate lipid synthesis in yeast. J. Biol. Chem. 287, 33364–33376 10.1074/jbc.M112.402339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Su W.-M., Han G.-S., and Carman G. M. (2014) Cross-talk phosphorylations by protein kinase C and Pho85p–Pho80p protein kinase regulate Pah1p phosphatidate phosphatase abundance in Saccharomyces cerevisiae. J. Biol. Chem. 289, 18818–18830 10.1074/jbc.M114.581462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hsieh L.-S., Su W.-M., Han G.-S., and Carman G. M. (2016) Phosphorylation of yeast Pah1 phosphatidate phosphatase by casein kinase II regulates its function in lipid metabolism. J. Biol. Chem. 291, 9974–9990 10.1074/jbc.M116.726588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Su W.-M., Han G.-S., and Carman G. M. (2014) Yeast Nem1–Spo7 protein phosphatase activity on Pah1 phosphatidate phosphatase is specific for the Pho85–Pho80 protein kinase phosphorylation sites. J. Biol. Chem. 289, 34699–34708 10.1074/jbc.M114.614883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Karanasios E., Han G.-S., Xu Z., Carman G. M., and Siniossoglou S. (2010) A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc. Natl. Acad. Sci. U.S.A. 107, 17539–17544 10.1073/pnas.1007974107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Karanasios E., Barbosa A. D., Sembongi H., Mari M., Han G.-S., Reggiori F., Carman G. M., and Siniossoglou S. (2013) Regulation of lipid droplet and membrane biogenesis by the acidic tail of the phosphatidate phosphatase Pah1p. Mol. Biol. Cell 24, 2124–2133 10.1091/mbc.e13-01-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pascual F., Hsieh L.-S., Soto-Cardalda A., and Carman G. M. (2014) Yeast Pah1p phosphatidate phosphatase is regulated by proteasome-mediated degradation. J. Biol. Chem. 289, 9811–9822 10.1074/jbc.M114.550103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hsieh L.-S., Su W.-M., Han G.-S., and Carman G. M. (2015) Phosphorylation regulates the ubiquitin-independent degradation of yeast Pah1 phosphatidate phosphatase by the 20S proteasome. J. Biol. Chem. 290, 11467–11478 10.1074/jbc.M115.648659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Siniossoglou S., Santos-Rosa H., Rappsilber J., Mann M., and Hurt E. (1998) A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17, 6449–6464 10.1093/emboj/17.22.6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang P.-C., Vancura A., Mitcheson T. G., and Kuret J. (1992) Two genes in Saccharomyces cerevisiae encode a membrane-bound form of casein kinase-1. Mol. Biol. Cell 3, 275–286 10.1091/mbc.3.3.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Robinson L. C., Hubbard E. J., Graves P. R., DePaoli-Roach A. A., Roach P. J., Kung C., Haas D. W., Hagedorn C. H., Goebl M., and Culbertson M. R. (1992) Yeast casein kinase I homologues: an essential gene pair. Proc. Natl. Acad. Sci. U.S.A. 89, 28–32 10.1073/pnas.89.1.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Snowdon C., and Johnston M. (2016) A novel role for yeast casein kinases in glucose sensing and signaling. Mol. Biol. Cell 27, 3369–3375 10.1091/mbc.E16-05-0342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Robinson L. C., Menold M. M., Garrett S., and Culbertson M. R. (1993) Casein kinase I-like protein kinases encoded by YCK1 and YCK2 are required for yeast morphogenesis. Mol. Cell. Biol. 13, 2870–2881 10.1128/MCB.13.5.2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stalder D., and Novick P. J. (2016) The casein kinases Yck1p and Yck2p act in the secretory pathway, in part, by regulating the Rab exchange factor Sec2p. Mol. Biol. Cell 27, 686–701 10.1091/mbc.E15-09-0651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., and Jensen O. N. (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4, 310–327 10.1074/mcp.M400219-MCP200 [DOI] [PubMed] [Google Scholar]
- 56. Li X., Gerber S. A., Rudner A. D., Beausoleil S. A., Haas W., Villén J., Elias J. E., and Gygi S. P. (2007) Large-scale phosphorylation analysis of α-factor–arrested Saccharomyces cerevisiae. J. Proteome Res. 6, 1190–1197 10.1021/pr060559j [DOI] [PubMed] [Google Scholar]
- 57. Chi A., Huttenhower C., Geer L. Y., Coon J. J., Syka J. E., Bai D. L., Shabanowitz J., Burke D. J., Troyanskaya O. G., and Hunt D. F. (2007) Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 104, 2193–2198 10.1073/pnas.0607084104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Smolka M. B., Albuquerque C. P., Chen S. H., and Zhou H. (2007) Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. U.S.A. 104, 10364–10369 10.1073/pnas.0701622104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Albuquerque C. P., Smolka M. B., Payne S. H., Bafna V., Eng J., and Zhou H. (2008) A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics 7, 1389–1396 10.1074/mcp.M700468-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Soufi B., Kelstrup C. D., Stoehr G., Fröhlich F., Walther T. C., and Olsen J. V. (2009) Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol. Biosyst. 5, 1337–1346 10.1039/b902256b [DOI] [PubMed] [Google Scholar]
- 61. Gnad F., de Godoy L. M., Cox J., Neuhauser N., Ren S., Olsen J. V., and Mann M. (2009) High-accuracy identification and bioinformatic analysis of in vivo protein phosphorylation sites in yeast. Proteomics 9, 4642–4652 10.1002/pmic.200900144 [DOI] [PubMed] [Google Scholar]
- 62. Helbig A. O., Rosati S., Pijnappel P. W., van Breukelen B., Timmers M. H., Mohammed S., Slijper M., and Heck A. J. (2010) Perturbation of the yeast N-acetyltransferase NatB induces elevation of protein phosphorylation levels. BMC Genomics 11, 685 10.1186/1471-2164-11-685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Soulard A., Cremonesi A., Moes S., Schütz F., Jenö P., and Hall M. N. (2010) The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell 21, 3475–3486 10.1091/mbc.e10-03-0182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bodenmiller B., Wanka S., Kraft C., Urban J., Campbell D., Pedrioli P. G., Gerrits B., Picotti P., Lam H., Vitek O., Brusniak M. Y., Roschitzki B., Zhang C., Shokat K. M., Schlapbach R., et al. (2010) Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci. Signal. 3, rs4 10.1126/scisignal.2001182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Swaney D. L., Beltrao P., Starita L., Guo A., Rush J., Fields S., Krogan N. J., and Villén J. (2013) Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 10, 676–682 10.1038/nmeth.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., McCartney R. R., Schmidt M. C., Rachidi N., Lee S. J., Mah A. S., et al. (2005) Global analysis of protein phosphorylation in yeast. Nature 438, 679–684 10.1038/nature04187 [DOI] [PubMed] [Google Scholar]
- 67. Vancura A., Sessler A., Leichus B., and Kuret J. (1994) A prenylation motif is required for plasma membrane localization and biochemical function of casein kinase I in budding yeast. J. Biol. Chem. 269, 19271–19278 [PubMed] [Google Scholar]
- 68. Babu P., Bryan J. D., Panek H. R., Jordan S. L., Forbrich B. M., Kelley S. C., Colvin R. T., and Robinson L. C. (2002) Plasma membrane localization of the Yck2p yeast casein kinase 1 isoform requires the C-terminal extension and secretory pathway function. J. Cell Sci. 115, 4957–4968 10.1242/jcs.00203 [DOI] [PubMed] [Google Scholar]
- 69. Herrador A., Livas D., Soletto L., Becuwe M., Léon S., and Vincent O. (2015) Casein kinase 1 controls the activation threshold of an α-arrestin by multisite phosphorylation of the interdomain hinge. Mol. Biol. Cell 26, 2128–2138 10.1091/mbc.E14-11-1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ingrell C. R., Miller M. L., Jensen O. N., and Blom N. (2007) NetPhosYeast: prediction of protein phosphorylation sites in yeast. Bioinformatics 23, 895–897 10.1093/bioinformatics/btm020 [DOI] [PubMed] [Google Scholar]
- 71. Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., and Brunak S. (2004) Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649 10.1002/pmic.200300771 [DOI] [PubMed] [Google Scholar]
- 72. Holt L. J., Tuch B. B., Villén J., Johnson A. D., Gygi S. P., and Morgan D. O. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 10.1126/science.1172867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lin Y.-P., and Carman G. M. (1990) Kinetic analysis of yeast phosphatidate phosphatase toward Triton X-100/phosphatidate mixed micelles. J. Biol. Chem. 265, 166–170 [PubMed] [Google Scholar]
- 74. Carman G. M., and Han G.-S. (2011) Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 80, 859–883 10.1146/annurev-biochem-060409-092229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pascual F., Soto-Cardalda A., and Carman G. M. (2013) PAH1-encoded phosphatidate phosphatase plays a role in the growth phase- and inositol-mediated regulation of lipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 288, 35781–35792 10.1074/jbc.M113.525766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., and O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- 77. Wang X., Hoekstra M. F., DeMaggio A. J., Dhillon N., Vancura A., Kuret J., Johnston G. C., and Singer R. A. (1996) Prenylated isoforms of yeast casein kinase I, including the novel Yck3p, suppress the gcs1 blockage of cell proliferation from stationary phase. Mol. Cell. Biol. 16, 5375–5385 10.1128/MCB.16.10.5375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Quon E., Sere Y. Y., Chauhan N., Johansen J., Sullivan D. P., Dittman J. S., Rice W. J., Chan R. B., Di Paolo G., Beh C. T., and Menon A. K. (2018) Endoplasmic reticulum-plasma membrane contact sites integrate sterol and phospholipid regulation. PLoS Biol. 16, e2003864 10.1371/journal.pbio.2003864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Harris T. E., Huffman T. A., Chi A., Shabanowitz J., Hunt D. F., Kumar A., and Lawrence J. C. Jr. (2007) Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J. Biol. Chem. 282, 277–286 10.1074/jbc.M609537200 [DOI] [PubMed] [Google Scholar]
- 80. Peterson T. R., Sengupta S. S., Harris T. E., Carmack A. E., Kang S. A., Balderas E., Guertin D. A., Madden K. L., Carpenter A. E., Finck B. N., and Sabatini D. M. (2011) mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146, 408–420 10.1016/j.cell.2011.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zanivan S., Gnad F., Wickström S. A., Geiger T., Macek B., Cox J., Fässler R., and Mann M. (2008) Solid tumor proteome and phosphoproteome analysis by high resolution mass spectrometry. J. Proteome Res. 7, 5314–5326 10.1021/pr800599n [DOI] [PubMed] [Google Scholar]
- 82. Grimsrud P. A., Carson J. J., Hebert A. S., Hubler S. L., Niemi N. M., Bailey D. J., Jochem A., Stapleton D. S., Keller M. P., Westphall M. S., Yandell B. S., Attie A. D., Coon J. J., and Pagliarini D. J. (2012) A quantitative map of the liver mitochondrial phosphoproteome reveals posttranslational control of ketogenesis. Cell Metab. 16, 672–683 10.1016/j.cmet.2012.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Humphrey S. J., Yang G., Yang P., Fazakerley D. J., Stöckli J., Yang J. Y., and James D. E. (2013) Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 17, 1009–1020 10.1016/j.cmet.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lundby A., Andersen M. N., Steffensen A. B., Horn H., Kelstrup C. D., Francavilla C., Jensen L. J., Schmitt N., Thomsen M. B., and Olsen J. V. (2013) In vivo phosphoproteomics analysis reveals the cardiac targets of β-adrenergic receptor signaling. Sci. Signal. 6, rs11 10.1126/scisignal.2003506 [DOI] [PubMed] [Google Scholar]
- 85. Huffman T. A., Mothe-Satney I., and Lawrence J. C. Jr. (2002) Insulin-stimulated phosphorylation of lipin mediated by the mammalian target of rapamycin. Proc. Natl. Acad. Sci. U.S.A. 99, 1047–1052 10.1073/pnas.022634399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Shimizu K., Fukushima H., Ogura K., Lien E. C., Nihira N. T., Zhang J., North B. J., Guo A., Nagashima K., Nakagawa T., Hoshikawa S., Watahiki A., Okabe K., Yamada A., Toker A., et al. (2017) The SCFβ-TRCP E3 ubiquitin ligase complex targets Lipin1 for ubiquitination and degradation to promote hepatic lipogenesis. Sci. Signal. 10, eaah4117 10.1126/scisignal.aah4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Eldar-Finkelman H., and Eisenstein M. (2009) Peptide inhibitors targeting protein kinases. Curr. Pharm. Des. 15, 2463–2470 10.2174/138161209788682253 [DOI] [PubMed] [Google Scholar]
- 88. Jenardhanan P., Panneerselvam M., and Mathur P. P. (2019) Targeting kinase interaction networks: a new paradigm in PPI based design of kinase inhibitors. Curr. Top. Med. Chem. 19, 467–485 10.2174/1568026619666190304155711 [DOI] [PubMed] [Google Scholar]
- 89. Wójcik P., and Berlicki L. (2016) Peptide-based inhibitors of protein–protein interactions. Bioorg. Med. Chem. Lett. 26, 707–713 10.1016/j.bmcl.2015.12.084 [DOI] [PubMed] [Google Scholar]
- 90. Jeffery D. A., Springer M., King D. S., and O'Shea E. K. (2001) Multi-site phosphorylation of Pho4 by the cyclin-CDK Pho80–Pho85 is semi-processive with site preference. J. Mol. Biol. 306, 997–1010 10.1006/jmbi.2000.4417 [DOI] [PubMed] [Google Scholar]
- 91. Sambrook J., Fritsch E. F., and Maniatis T. (1989) Molecular Cloning, A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 92. Innis M. A., and Gelfand D. H. (1990) in PCR Protocols. A Guide to Methods and Applications (Innis M. A., Gelfand D. H., Sninsky J. J., and White T. J., eds) pp. 3–12, Academic Press, Inc., San Diego [Google Scholar]
- 93. Han G.-S., Sreenivas A., Choi M.-G., Chang Y.-F., Martin S. S., Baldwin E. P., and Carman G. M. (2005) Expression of human CTP synthetase in Saccharomyces cerevisiae reveals phosphorylation by protein kinase A. J. Biol. Chem. 280, 38328–38336 10.1074/jbc.M509622200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Rose M. D., Winston F., and Heiter P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 95. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 96. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 97. Carman G. M., and Matas J. (1981) Solubilization of microsomal-associated phosphatidylserine synthase and phosphatidylinositol synthase from Saccharomyces cerevisiae. Can. J. Microbiol. 27, 1140–1149 10.1139/m81-179 [DOI] [PubMed] [Google Scholar]
- 98. Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., and Séraphin B. (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 10.1006/meth.2001.1183 [DOI] [PubMed] [Google Scholar]
- 99. Gerace E., and Moazed D. (2015) Affinity purification of protein complexes using TAP tags. Methods Enzymol. 559, 37–52 10.1016/bs.mie.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Boyle W. J., van der Geer P., and Hunter T. (1991) Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201, 110–149 10.1016/0076-6879(91)01013-R [DOI] [PubMed] [Google Scholar]
- 101. Yang W.-L., and Carman G. M. (1995) Phosphorylation of CTP synthetase from Saccharomyces cerevisiae by protein kinase C. J. Biol. Chem. 270, 14983–14988 10.1074/jbc.270.25.14983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. MacDonald J. I., and Kent C. (1994) Identification of phosphorylation sites in rat liver CTP:phosphocholine cytidylyltransferase. J. Biol. Chem. 269, 10529–10537 [PubMed] [Google Scholar]
- 103. Guengerich F. P., Wang P., and Davidson N. K. (1982) Estimation of isozymes of microsomal cytochrome P-450 in rats, rabbits, and humans using immunochemical staining coupled with sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochemistry 21, 1698–1706 10.1021/bi00536a035 [DOI] [PubMed] [Google Scholar]
- 104. Burnette W. (1981) Western blotting: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112, 195–203 10.1016/0003-2697(81)90281-5 [DOI] [PubMed] [Google Scholar]
- 105. Haid A., and Suissa M. (1983) Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 96, 192–205 10.1016/S0076-6879(83)96017-2 [DOI] [PubMed] [Google Scholar]
- 106. Havriluk T., Lozy F., Siniossoglou S., and Carman G. M. (2008) Colorimetric determination of pure Mg2+-dependent phosphatidate phosphatase activity. Anal. Biochem. 373, 392–394 10.1016/j.ab.2007.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Henry S. A., Kohlwein S. D., and Carman G. M. (2012) Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190, 317–349 10.1534/genetics.111.130286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Park Y., Han G. S., and Carman G. M. (2017) A conserved tryptophan within the WRDPLVDID domain of yeast Pah1 phosphatidate phosphatase is required for its in vivo function in lipid metabolism. J. Biol. Chem. 292, 19580–19589 10.1074/jbc.M117.819375 [DOI] [PMC free article] [PubMed] [Google Scholar]


