Abstract
Heterochromatin is a conserved feature of eukaryotic genomes and regulates various cellular processes, including gene silencing, chromosome segregation, and maintenance of genome stability. In the fission yeast Schizosaccharomyces pombe, heterochromatin formation involves methylation of lysine 9 in histone H3 (H3K9), which recruits Swi6/HP1 proteins to heterochromatic loci. The Swi6/HP1–H3K9me3 chromatin complex lies at the center of heterochromatic macromolecular assemblies and mediates many functions of heterochromatin by recruiting a diverse set of regulators. However, additional factors may be required for proper heterochromatin organization, but they are not fully known. Here, using several molecular and biochemical approaches, we report that Vgl1, a member of a large family of multiple KH-domain proteins, collectively known as vigilins, is indispensable for the heterochromatin-mediated gene silencing in S. pombe. ChIP analysis revealed that Vgl1 binds to pericentromeric heterochromatin in an RNA-dependent manner and that Vgl1 deletion leads to loss of H3K9 methylation and Swi6 recruitment to centromeric and telomeric heterochromatic loci. Furthermore, we show that Vgl1 interacts with the H3K9 methyltransferase, Clr4, and that loss of Vgl1 impairs Clr4 recruitment to heterochromatic regions of the genome. These findings uncover a novel role for Vgl1 as a key regulator in heterochromatin-mediated gene silencing in S. pombe.
Keywords: chromatin, chromatin modification, gene silencing, histone methylation, heterochromatin
Introduction
The eukaryotic genome is spatially segregated in the nucleus into discrete structural and functional chromatin domains (1–5). The genome is organized into euchromatin and heterochromatin distinguished on the basis of their appearance, organization, localization, function, and the type of histone modifications on nucleosomes (6, 7). Heterochromatin is a typically condensed chromatin structure refractory to transcriptional machinery and is important for various cellular processes, including regulation of gene expression, chromosome segregation, dosage compensation, and maintenance of genome stability by inhibiting unwanted recombination between repetitive DNA elements (8, 9). Across species, assembly of heterochromatin involves coordinating activities of small noncoding RNAs associated with the RNA interference (RNAi) pathway and chromatin-modifying machinery (10–13). In fission yeast, Schizosaccharomyces pombe heterochromatin is found at the telomeres, the mating-type and ribosomal DNA loci, and the pericentromeric region. The centromeres are composed of a central core region, which is excluded from heterochromatin-specific factors, and modifications flanked by innermost repeat (imr)3 and outer (otr) repeat, called dg and dh repeats (14–19). These repeats are transcribed into dsRNAs by RNA pol II and are further processed into siRNAs by RNAi factors, including Argonaute (Ago1), Dicer (Dcr1), and RNA-dependent RNA polymerase (Rdp1). RNA transcripts act as a scaffold for the assembly of RNAi and chromatin-modifying factors that initiate the formation of heterochromatin (20–23). Deletion of fission yeast RNAi proteins like Dcr1, Rdp1, and Argonaute results in loss of heterochromatin gene silencing (24–27). Histone H3 lysine 9 methylation (H3K9me), mediated by a conserved Clr4 methyltransferase, is specifically localized in heterochromatic regions of the genome. H3K9 methylation creates binding sites for HP1 proteins Swi6 and Chp2, which are critical for heterochromatic gene silencing (28–36). Nucleation and spreading of H3K9 methylation depends on two distinct processes: an RNAi-independent pathway mediates low levels of H3K9me, and an RNAi-dependent mechanism boosts this methylation. In the absence of RNAi, H3K9me fails to spread, and silencing of reporter genes inserted within the centromeres is abrogated (28, 37, 38). In addition to RNAi, H3K9me, and Swi6, other additional factors have also been associated with heterochromatic gene silencing (39, 40). Recently, we have reported that Lem2–Nur1 inner nuclear membrane complex is essential for heterochromatin gene silencing (41). Tandem affinity purification of Lem2 or Nur1 identified Vgl1, a predicted heterochromatin protein, as an interacting partner. Vgl1 is an evolutionarily-conserved protein from the yeast Saccharomyces cerevisiae (Scp160) to Drosophila (DDP1) and vertebrates (Vigilin) and belongs to a large family of multiple KH-domain proteins, collectively known as vigilins (42, 43). KH domains are involved in protein–protein and protein–nucleic acid interactions. Several KH domain-containing proteins bind RNA and are involved in RNA transport and metabolism (44). In the Drosophila vigilin homologue, DDP1 localizes to heterochromatin, and depletion of DDP1 results in changed nuclear morphology and defects in heterochromatin due to aberrant pericentromeric heterochromatin formation. DDP1-deleted cells show that suppression of the heterochromatin-induced position affects variegation and mislocalization of HP1 (45–48). In S. cerevisiae Scp160p, which is a vigilin homologue, has been shown to be important for chromosome segregation as deletion causes chromosome segregation defects (49–51). However, the role of Vgl1 in heterochromatin gene silencing in S. pombe is not known. Here, we report that Vgl1 is essential for heterochromatin integrity in S. pombe. Our data show that Vgl1 interacts with H3K9 methyltransferase, Clr4, and deletion of Vgl1 impairs the recruitment and activity of heterochromatin machinery. Taken together, our findings reveal a previously unknown role of Vgl1 as an essential component in heterochromatic gene silencing in S. pombe.
Results
Vgl1 Is Required for Heterochromatin Integrity
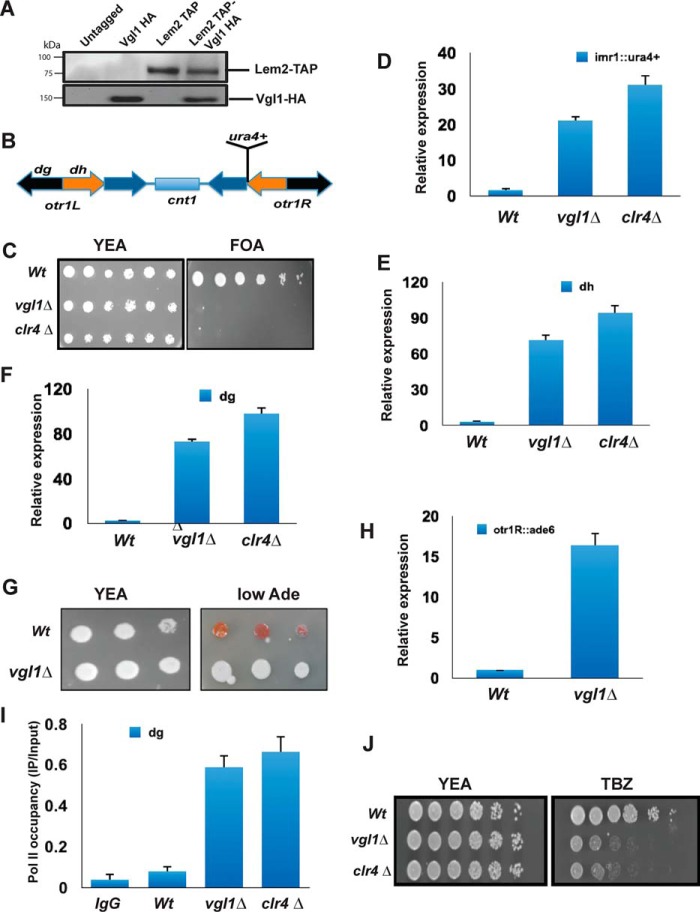
Recently, we have identified an inner nuclear membrane protein complex Lem2–Nur1 as important for heterochromatin gene silencing and its localization toward nuclear periphery (40, 41). Tandem affinity purifications of Lem2 or Nur1 identified Vgl1 as an interacting partner (Fig. 1A). To gain insights about the role of the Vgl1 in heterochromatin gene silencing, we deleted vgl1 in a parental strain in which the native ura4+ gene was deleted, and the ura4+ reporter gene was inserted at centromeric otr1R region (Fig. 1B). The silencing of ura4+ by heterochromatin enables cells to grow on media containing 5-fluoroorotic acid (FOA), a drug toxic to cells that express ura4+. As shown in Fig. 1C, deletion of vgl1 leads to loss of silencing of the otr1R::ura4+ reporter as determined by growth on media containing FOA. Quantitative RT-PCR analysis of the otr1R::ura4+ reporter or endogenous transcripts originating from the dg and dh elements of centromeres confirmed the silencing phenotypes observed in vgl1-deleted cells (Fig. 1, D–F). We used clr4Δ as a positive control where heterochromatin is completely disrupted (20, 28). We further examined the stability of heterochromatin upon vgl1 depletion in cells having ade6+ reporter gene integrated at centromere (otr1:: ade6+). The silencing of ade6+ results in the formation of red colonies when cells are grown in low-adenine medium and disruption of heterochromatin results in the formation of white colonies. On low Ade plates, vgl1 loss leads to formation of white colonies, suggesting disruption of heterochromatin (Fig. 1G). ade6+ reporter gene derepression was verified by quantitative RT-PCR (qRT-PCR) to examine Ade6+ transcript levels (Fig. 1H). Consistent with plating assays and quantitative RT-PCR results, polymerase II occupancy at centromeres increases in vgl1-deleted cells (Fig. 1I). Heterochromatin integrity at centromeres is essential for faithful chromosome segregation, and loss of centromeric heterochromatin causes defects in chromosome segregation during mitosis and exhibits increased incidence of lagging chromosomes and hypersensitivity to the microtubule-destabilizing agent thiabendazole (TBZ) (52). The segregation defect is due to incorrect sister chromatid orientation and the defective attachment of the kinetochore to the spindle (20, 53, 54). We observed that cells lacking vgl1 showed hypersensitivity to the microtubule-destabilizing drug TBZ (Fig. 1J). These results are consistent with the role of Scp160p in S. cerevisiae, where its deletion results in ploidy as well as in Drosophila and humans, where knockdown of vigilin causes defects in chromosome condensation and chromosome lagging during cell division. The overall results indicate that Vgl1 is essential for heterochromatic gene silencing at pericentromeric heterochromatin and for normal functioning of centromeres.
Figure 1.
Vgl1 is essential for heterochromatin integrity. A, coimmunoprecipitation experiments followed by Western blot analysis showing interaction between Lem2 and Vgl1. Cells expressing Lem2–TAP and Vgl1–HA were lysed, and Lem2–TAP was pulled down using anti-TAP antibody. Immunoprecipitated fractions were analyzed by Western blotting using anti-HA antibodies. B, schematic diagram of centromere1 of S. pombe depicting central core region (cnt1), imr, and otr containing dg and dh repeats. Position of the cen1:ura+ marker gene at imr1R is also shown in the diagram. C, 10-fold serial dilution plating assays of WT (WT) and vgl1Δ were used to observe gene silencing of imr1R::ura4 on counter-selective FOA medium; clr4Δ strain was used as a positive control. D–F, qRT-PCR analysis of cen dg, dh, and reporter imr::ura+ transcript levels relative to a control transcript act1+. G, 10-fold serial dilution plating assays used to examine gene silencing of otr::ade6+ in WT and Vgl1-deleted cells. WT cells form red colonies on limiting adenine plates, whereas loss of heterochromatin silencing leads to white colonies. H, qRT-PCR analysis of ade6+ transcript levels in the indicated strains. The error bars represent standard deviation from three independent experiments, and all the values were normalized to act1. I, ChIP analysis of pol II occupancy at dg repeats in WT and vgl1Δ background with clr4Δ as a positive control. J, plating assay showing sensitivity of vgl1Δ cells to the microtubule-destabilizing drug TBZ.
Vgl1 binds to pericentromeric heterochromatin and regulates H3K9 methylation and Swi6 recruitment
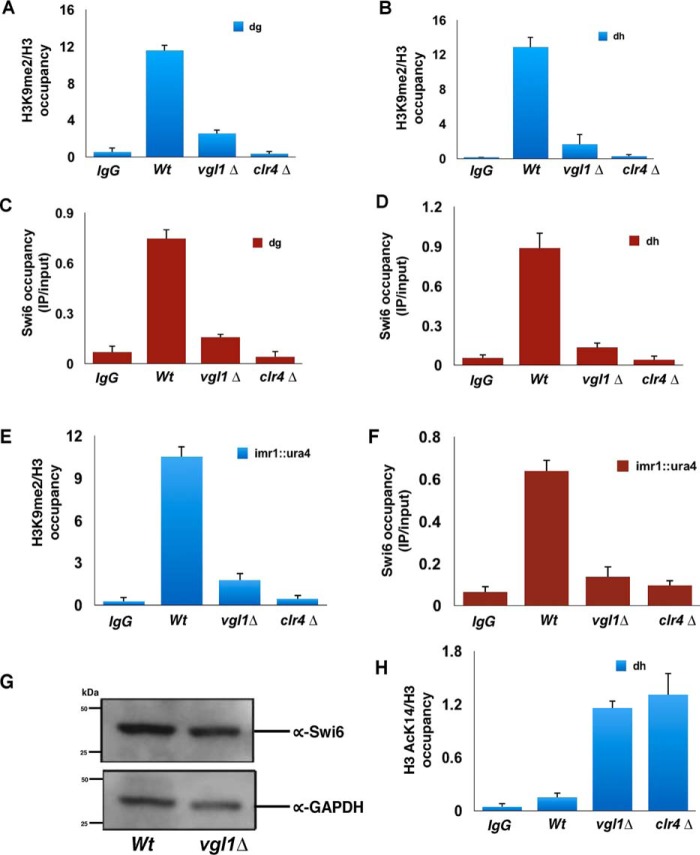
To gain further insight into the function of Vgl1 in heterochromatin organization and to see whether the silencing by Vgl1 is direct, we generated strains expressing functional C-terminally HA-tagged Vgl. ChIP analysis of Vgl1 showed that it binds to pericentromeric regions (Fig. 2, A and B). Furthermore, the HA-tagged Vgl1 strain was functional as confirmed by dilution assay (Fig. 2C). One of the key steps in S. pombe heterochromatin formation involves methylation of H3K9 by Clr4 methyltransferase, which is a hallmark of constitutive heterochromatin in most eukaryotes. This methylation acts as a landing platform for heterochromatin protein Swi6, which is important for heterochromatin formation and spreading (28–36). To gain more insights about the molecular mechanism of how Vgl1 regulates heterochromatin gene silencing, we looked at the impact of vgl1 deletion on H3K9 methylation and Swi6 enrichment at centromeric heterochromatin. ChIP against H3K9me2 in cells lacking Vgl1 shows complete loss of H3K9 methylation from heterochromatic regions (Fig. 3, A and B). Consistent with H3K9 methylation data, Swi6 levels in vgl1-deleted cells were abolished at native heterochromatic regions (dg and dh, Fig. 3, C and D). In addition, ChIP analysis showed that like native heterochromatic regions, cells lacking vgl1 showed reduced levels of H3K9 methylation and Swi6 enrichment over the reporter gene (otr1R::ura4+, Fig. 3, E and F) indicating a defect in the integrity of pericentromeric heterochromatin. Furthermore, H3K9 methylation and Swi6 loss in vgl1-deleted cells is comparable with that of clr4Δ cells. In addition, decreased levels of Swi6 binding to chromatin in cells lacking Vgl1 are not due to decreased Swi6 protein levels (Fig. 3G). To further test whether loss of vgl1 impacts modifications associated with transcriptional activation at pericentromeric heterochromatin, we measured H3K14 acetylation levels in WT and vgl1-deleted cells by ChIP. Consistent with a drop in H3K9 methylation and Swi6 recruitment, we observed that the acetylation (H3K14ac) levels show a 5-fold increase in vgl1 mutant cells compared with that in the WT at pericentric regions (Fig. 3H). These findings suggest that Vgl1 regulates pericentromeric gene silencing directly by associating with the centromeric heterochromatin and is required to maintain integrity and repressive chromatin modifications at centromeric heterochromatin. These results are consistent with the Drosophila homologue DDP1, which has been previously shown to colocalize with HP1, and the loss of DDP1 showed strong reduction in H3K9 methylation (45–48).
Figure 2.
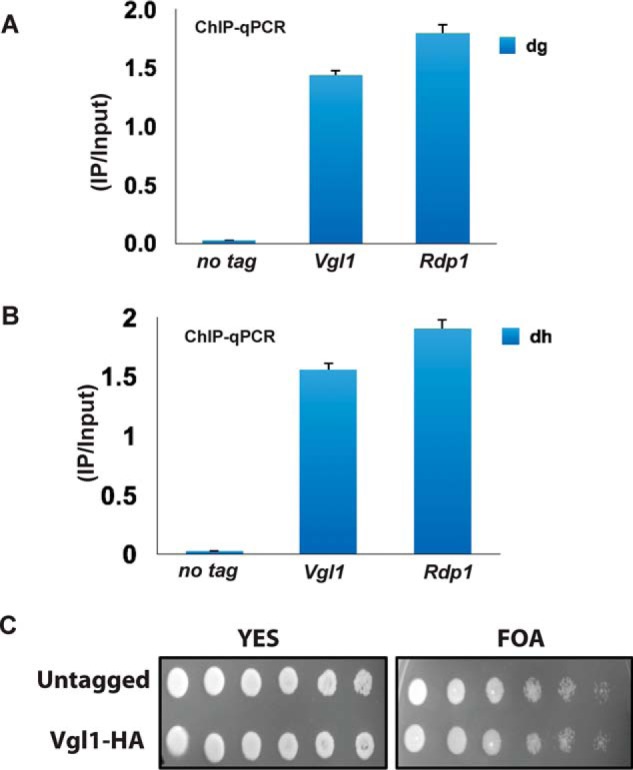
Vgl1 binds to pericentromeric heterochromatin regions. A and B, ChIP experiments showing the binding of Vgl1 to pericentromeric dg and dh repeats. Rdp1 was used as a positive control. C, serial dilution assay showing the functionality of tagged Vgl1.
Figure 3.
Vgl1 regulates H3K9 methylation and Swi6 recruitment to pericentromeric heterochromatin. A–D, ChIP experiments showing that loss of Vgl1 causes decrease in H3K9me2 and Swi6 binding at the centromeric dg and dh repeats. E and F, H3K9me2 levels and Swi6 recruitment was also dramatically reduced at the imr1::ura4 reporter gene in Vgl1-deleted cells. G, Western blotting of Swi6 protein levels in WT and vgl1Δ cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. H, ChIP analysis showing increased H3K14Ac levels upon Vgl deletion at dg repeats. Clr4Δ and Swi6Δ strains were used as a control in all the ChIP experiment, and error bars represent standard deviations from three independent experiments.
Vgl1 regulates Clr4 binding to pericentromeric heterochromatin
Loss of H3K9 methylation and Swi6 recruitment to chromatin in vgl1Δ cells allowed us to speculate whether Vgl1 plays a role in recruitment of H3K9 methyltransferase, Clr4, to the pericentromeric heterochromatin and whether loss of vgl1 impairs Clr4 recruitment to heterochromatin. To test this, we deleted vgl1 in a strain expressing N-terminally FLAG-tagged Clr4 and looked at the binding of Clr4 in vgl1-deleted cells by ChIP. Interestingly, ChIP experiments show that FLAG–Clr4 association with centromeres was reduced to background levels in vgl1-deleted cells (Fig. 4, A and B). Furthermore, immunoblot analysis showed that decreased binding of Clr4 to heterochromatin in Vgl1-depleted cells is not due to decreased levels of Clr4 protein levels, which are similar to WT cells (Fig. 4C). These results suggest that Vgl1 controls H3K9 methylation by directly regulating the recruitment of Clr4 to the centromeric heterochromatin.
Figure 4.
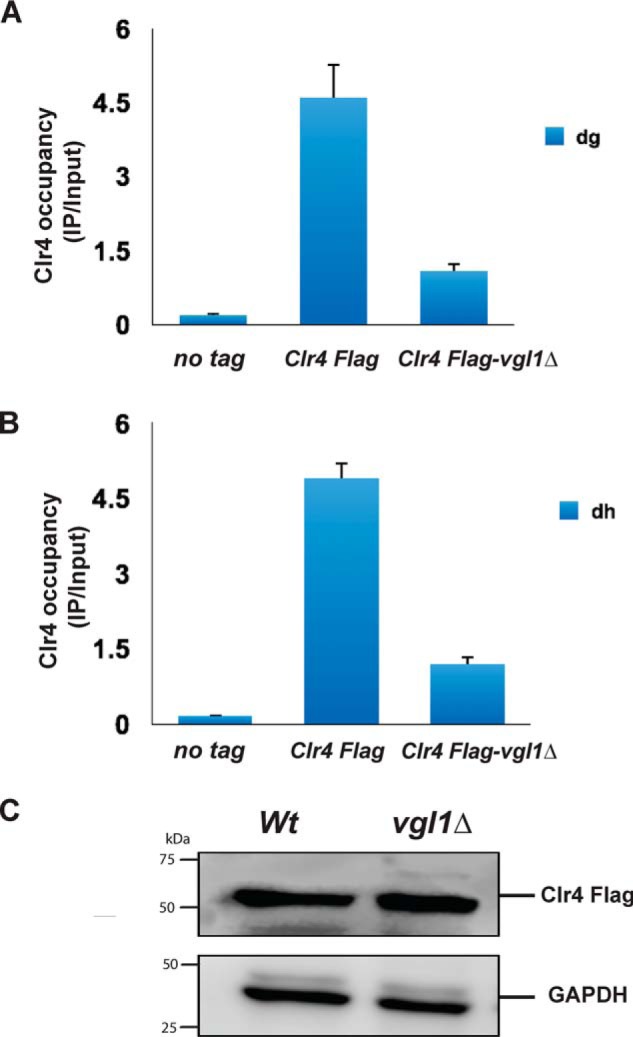
Vgl1 regulates Clr4 binding to pericentromeric heterochromatin. A and B, ChIP–qPCR analysis of Clr4–FLAG binding to pericentromeric heterochromatin in the indicated strains. C, immunoblot of Clr4 protein levels in WT and vgl1Δ cells indicating that impairment of Clr4 recruitment to chromatin in Vgl1-deleted cells is not due to change in Clr4 protein levels.
Vgl1 is required for integrity of telomeric heterochromatin
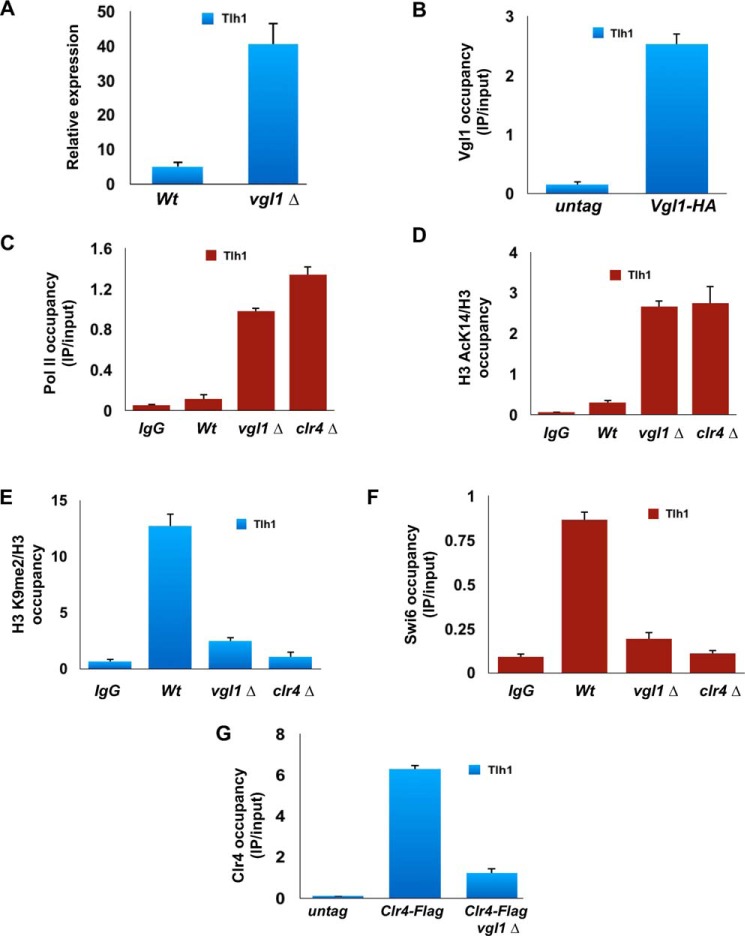
Apart from centromeres, heterochromatin domains in S. pombe are also found at telomeres, where an alternative pathway independent of RNAi works for establishment of heterochromatin. To gain further insights about the role of Vgl1 in telomeric heterochromatin organization, we looked at the expression of the subtelomeric gene, tlh1, in WT and vgl1-deleted cells, as shown in Fig. 5A, and loss of vgl1 causes derepression of the tlh1 gene. Furthermore, ChIP analysis of Vgl1 showed that it binds to telomeric heterochromatin, and in tune with silencing defect at telomeres, vgl1-deficient cells show increased polymerase II occupancy and higher levels of histone acetylation at telomeric heterochromatin (Fig. 5, B–D). In addition, cells lacking vgl1 display reduced levels of H3K9 methylation and Swi6 enrichment to telomeric heterochromatin loci (Fig. 5, E and F). Like centromeric heterochromatin, loss of Vgl1 impairs recruitment of Clr4 to telomeric regions of the genome (Fig. 5G). These results suggest that Vgl1 is essential for maintaining the silencing of telomeric heterochromatin in the same pathway as at centromeres.
Figure 5.
Vgl1 regulates telomeric heterochromatin. A, qRT-PCR analysis of Tlh1 transcript levels relative to a control transcript act1+ in WT and vgl1-deleted cells. B–D, ChIP analysis showing binding of Vgl1, pol II enrichment, and H3K14 acetyl levels at telomeric heterochromatin in WT and vgl1-deficient cells. E and F, H3K9me2 levels and Swi6 recruitment were significantly reduced at telomeres in Vgl1-deleted cells. G, ChIP–qPCR analysis of Clr4–FLAG binding to telomeric heterochromatin in WT and vgl1-deleted cells.
Vgl1 interacts with H3K9 histone methyltransferase Clr4
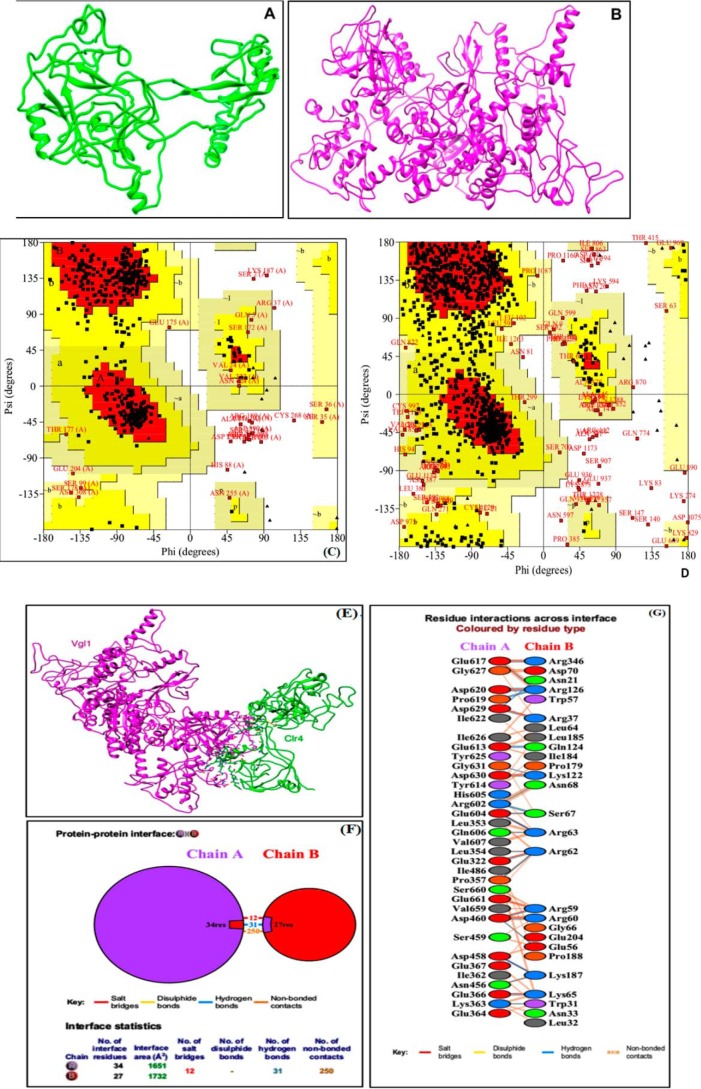
To gain further insights into the regulation of H3K9 methylation and recruitment of Clr4 to pericentromeric heterochromatin by Vgl1, we carried out protein–protein docking studies (Fig. 6, A–D), followed by extensive interaction profile generation, which suggest that Vgl1 and Clr4 interact through various interactions, including hydrogen bonding, salt bridges, and other nonbonded contacts. Our protein–protein interface analysis indicated that 34 amino acid residues of Vgl1 and 27 residues of Clr4 interact with each other, forming 12 salt bridges, 31 hydrogen bonds, and 250 nonbonded contacts (Fig. 6, E–G). Vgl1 has 12 KH domains ranging from KH1 to KH12 (42, 43), whereas Clr4 possesses chromo-domain and SET domains (28–36). Our in silico predictions suggest that chromo-domain of Clr4 and the KH5 domain of Vgl1 are the most critical for interaction. This conclusion is drawn from the profile of residue interactions across the interface where out of the 27 interacting residues of Clr4, 16 belong to chromo-domain (residues 8–69 of chain B) and one amino acid residue, Arg-346, belongs to its SET domain (residues 328–452 of B chain). Regarding Vgl1 among the 35 interacting residues, 17 belong to the KH5 domain (residues 575–644 of chain A), seven to the KH3 domain (339–405 of chain A), three to the KH4 (416–486) and KH6 domains (658–726), and one (Glu-322) to the KH2 domain (236–328 of chain A) Fig. 6, F and G.
Figure 6.
Molecular docking of Vgl1 and Clr4. A and B, refined models of Clr4 and Vgl1. The respective sequences of Vgl1 and Clr4 proteins were retrieved from Universal Protein Resource (UniPROT), and then models were generated using the futuristic software I-TASSER. As a rule of thumb, the crude models were refined using the GalaxyRefine and ModRefiner, respectively. GalaxyRefine improves both the local and global quality of protein models. ModRefiner substantially improves the physical quality of local structures and is regarded as a high-resolution protein structure refinement algorithm. C, stereochemical quality of refined Clr4 model was checked by Ramachandran plot. This plot was generated using PROCHECK of Structure Analysis and Verification Server (SAVES)-server. This plot provides insights not only regarding residue by residue geometry but also about the overall structure geometry. Refined model of Clr4 showed 77.8% residues in most favored regions, 15.8% residues in additionally-allowed regions, 3.2% residues in generously-allowed regions, and only 3.4% residues in disallowed regions. D, Ramachandran plot of refined Vgl1 generated through PROCHECK. The refined model showed 68.6% residues in most favored regions, 24.5% residues in additionally-allowed regions, 4.4% residues in generously-allowed regions, and only 2.4% residues in disallowed regions. E, docked complex of Vgl1 and Clr4 showing the interaction of these proteins. Vgl1 (chain A) and Clr4 (chain B) interact via multiple interactions like hydrogen bonding, salt bridges, and other nonbonded contacts. The statistics of the protein–protein interface diagram clearly shows that 34 residues from Vgl1 side interact with 24 residues of Clr4. The two proteins interact strongly as evidenced by 12 salt bridges, 31 hydrogen bonds, and 250 nonbonded contacts. F, KH5 domain (575–644 amino acid residues) of Vgl1 (chain A) is most crucial for interaction as 17 residues among the 35 interacting residues belong to this domain followed by seven residues of KH3 domain (339–405), three residues of KH4(416–486), three residues of KH6(658–726), and one residue of the KH2 domain (236–328). Regarding Clr4, the chromo-domain has a key role in interaction as 16 among 27 interacting residues belong to this domain (residues 8–69). Moreover, one residue of Clr4 (Arg-346) from the SET domain (328–452) also interacts with Vgl1. G, refer to keys provided with F and G and residue colors provided below G for finer details.
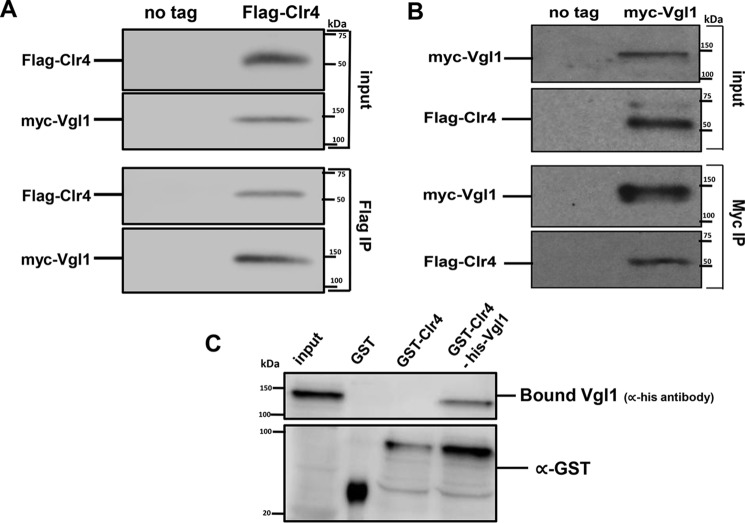
To validate our protein–protein docking studies, we used coimmunoprecipitation assays to test whether Vgl1 physically associates with the H3K9 histone methyltransferase Clr4. We performed immunoprecipitation experiments from cells that expressed functional C-terminally HA-tagged Vgl1 and N-terminally Myc-tagged Clr4 expressed under the control of their endogenous promoters. As shown in Fig. 7A, immunoprecipitation with anti-Myc antibody followed by Western blotting with anti-HA antibody showed that Vgl1 associates with Clr4. Reciprocal immunoprecipitation with anti-HA antibody followed by Western blotting with anti-Myc antibody conformed the physical interaction between Clr4 and Vgl1 (Fig. 7B). To further validate our in silico and coimmunoprecipitation data and to test whether Vgl1 directly interacts with Clr4, we performed GST pulldown assays with recombinant GST–Clr4 and His tagged Vgl1. As shown in Fig. 7C, Vgl1 efficiently binds to Clr4 in vitro. These results show that Vgl1 regulates H3K9 methylation by directly interacting with Clr4 and acts as a key protein that connects the CLRC complex to centromeric heterochromatin.
Figure 7.
Vgl1 interacts with H3K9 histone methyltransferase Clr4 in vivo. A and B, Western blot analysis of coimmunoprecipitation experiments shows that myc–Clr4 immunoprecipitates Vgl1–HA. Cell extracts prepared from myc–Clr4 and HA–Vgl1 or from untagged strains were incubated with anti-Myc antibody, and immunoprecipitated fractions were analyzed by Western blotting with anti-HA antibodies. B, HA antibody was used for immunoprecipitation, and immunoprecipitated fractions were analyzed by Western blotting with anti-Myc antibodies. C, GST pulldown assay showing the association of Clr4 and the Vgl1. GST is used as a negative control. After the washes, input and bound fractions were run on a 10% SDS-PAGE followed by Western blotting using anti-His and anti-GST antibodies.
Vgl1 relies on direct RNA binding for recruitment to centromeric heterochromatin
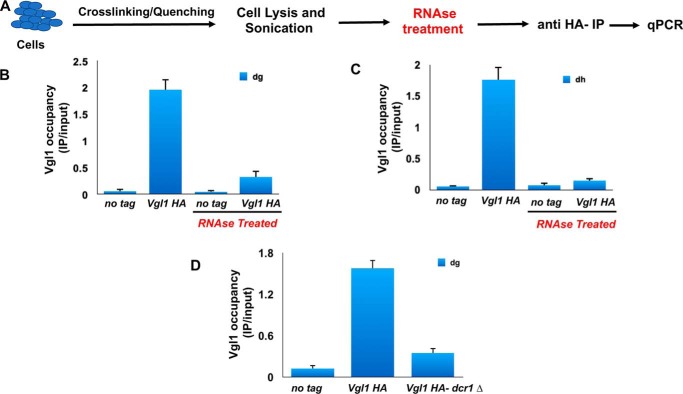
The Vgl1/Vigilin family of proteins contain multiple tandemly arranged KH domains. KH domains are one of the best-characterized family of RNA-binding proteins. We next examined whether the binding of Vgl1 to heterochromatin is RNA-dependent. To specifically test the contribution of RNA to Vgl1 localization to pericentromeric heterochromatin, we modified the ChIP protocol slightly by treating the cell lysate with RNase for 30 min before immunoprecipitation (Fig. 8A). We found that RNase treatment significantly reduced the Vgl1 recruitment to heterochromatic regions compared with undigested control cells (Fig. 8, B and C). To gain further insights about the functional relationship between Vgl1 and RNAi machinery, we carried out ChIP analysis to look at the binding of Vgl1 to centromeric heterochromatin in WT and dcr1Δ cells. Interestingly, like the RNase data, association of Vgl1 with chromatin was significantly reduced in dcr1Δ cells (Fig. 8D). These results suggest that like other RNAi-directed chromatin components, Vgl1 association with centromeric heterochromatin depends on the siRNA pathway and suggests that RNAi components are required for Vgl1 association with heterochromatin.
Figure 8.
Association of Vgl1 with chromatin is RNase-sensitive. A, schematic of ChIP with an RNase step. B and C, ChIP–qPCR analysis of HA-Vgl1 binding to dg and dh centromeric repeats in the indicated strains either untreated or treated with RNase. D, ChIP analysis showing binding of Vgl1 to centromeric heterochromatin in WT and dcr1-deficient cells.
Discussion
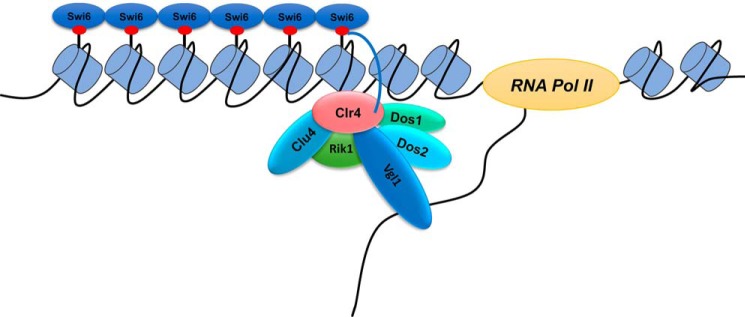
Chromatin within the cell nucleus is segregated into euchromatin and heterochromatin distinguished on the basis of their appearance, structure, localization, and function. This spatiotemporal distribution of chromatin domains correlates with their functional state (6–9). In fission yeast, S. pombe heterochromatin domains are found at the telomeres, mating-type and ribosomal DNA loci, and the pericentromeric region. Establishment of proper heterochromatin at centromeres is critical for chromosome segregation as it provides a binding platform for kinetochore assembly (14–19). The assembly of heterochromatin relies on a complex interplay between RNAi factors, chromatin-modifying proteins, and nonhistone proteins that are important for the establishment of heterochromatin and for its maintenance (20–23, 39, 40). However, there is a significant gap in our understanding of the full array of nonhistone proteins that coordinate with chromatin-modifying and RNAi machinery in the heterochromatin organization in S. pombe. In this study, we have provided the first characterization of a novel gene, Vgl1, with a pivotal role in heterochromatic gene-silencing in S. pombe. Whereas Vgl1 is known to play an essential role in various cellular processes like heterochromatin formation in Drosophila, chromosome segregation in S. cerevisiae, mRNA stability, and regulation of the imprinted genes Igf2 and H19 through its association with the CCCTC-binding factor (CTCF) (42–51). This is the first time Vgl1 has been shown to be essential for heterochromatin formation in S. pombe. Our results show that loss of vgl1 leads to heterochromatin derepression at centromeres and integrated ura4+ and Ade6+ reporter genes. The silencing phenotypes are comparable with that of clr4 deletion (Fig. 1). Vgl1 mutant cells also show segregation defects due to instability of heterochromatin at centromeres. These results are consistent with the role of vgl1 in Drosophila, where knockdown of DDP1 causes a heterochromatin defect due to aberrant pericentromeric heterochromatin formation. Similarly, deletion of Scp160 in S. cerevisiae causes segregation defects (45–51). Furthermore, our results show that Vgl1 binds to pericentomeric and telomeric heterochromatin and regulates H3K9 methylation and Swi6 recruitment (Figs. 2 and 5). We show that the silencing defect at centromeres and telomeres is due to decreased H3K9 methylation levels, reduced Swi6 recruitment, and increased levels of H3K14 acetylation (Figs. 3 and 5). On the molecular level, regulation of heterochromatin by Vgl1 involves binding to a nascent transcript and subsequent recruitment of the CLRC complex to chromatin, which methylates H3K9 and recruits Swi6 resulting in transcriptional gene silencing and heterochromatin spreading. Our studies suggest that Vgl1 provides a critical link between siRNA and Clr4–H3K9 methylation, a pathway that lies at the center of heterochromatic macromolecular assemblies and mediates many functions of heterochromatin. Our conclusions are based on the following: first, Vgl1 deletion impairs Clr4 recruitment to chromatin and results in loss of H3K9 methylation at centromeres, reporter genes; second, Vgl1 associates with Clr4 in vivo and is crucial for its recruitment to modify local chromatin for proper heterochromatin formation; and third, RNase treatment or deletion of Dcr1 abolishes Vgl1 recruitment to chromatin. Based on these findings, we propose that Vgl1 binds to centromeric transcripts and either helps in CLRC recruitment or stabilizes its association with chromatin (Fig. 9). Vgl1 appears to be an adaptor protein and mediates CLRC interaction with chromatin. In summary, our findings uncover a central role for Vgl1, a highly-conserved protein from yeast to humans, as an essential factor critical for heterochromatic silencing in S. pombe. Our study demonstrates for the first time that the multiple KH-domain protein Vgl1 plays a critical role in heterochromatin regulation in S. pombe. In the future, it would be intriguing to investigate whether Vgl1 plays a role in stability of repetitive DNA elements across species and whether Vgl1 has a role in higher-order genome organization.
Figure 9.
Model showing heterochromatin gene silencing by Vgl1. Vgl1 gets recruited to pericentromeric heterochromatin by interacting with the nascent transcript and either helps in recruitment of Clr4 to chromatin or stabilizes its association with pericentromeric heterochromatin. Clr4 leads to the methylation of H3K9 and subsequent recruitment of Swi6.
Materials and methods
Yeast strains
All the strains used in this study are given in Table 1. The strains were generated by PCR-based gene targeting method wherein the primers contain sequences homologous to the flanking sequences of targeted sites for efficient recombination (55), and positive clones were selected on the basis of growth on YEA plates containing the desired antibiotic and were further confirmed by PCR.
Table 1.
The list of strains used in this study
| Strain no. | Strain | Genotype | Source |
|---|---|---|---|
| 1. | SPY137 | h + otr1R(SphI)::ura4 + ura4-DS/E leu1–32 ade6-M210 | 41 |
| 2. | SPY815 | h + otr1R(SphI)::ura4 + ura4-DS/E leu1–32 ade6-M210 clr4Δ::KanMX | 41 |
| 3. | AB099 | h + otr1R(SphI)::ura4 + ura4-DS/E leu1–32 ade6-M210 vgl1Δ::KanMX | This study |
| 4. | SPY816 | h + otr1R(SphI)::ura4 + ura4-DS/E leu1–32 ade6-M210 swi6Δ::KanMX | 41 |
| 5. | FY511 | h90 mat3-M::ura4 ade6–216 leu 1–32 ura4-D18 | 39 |
| 6. | AB141 | h + ade6–210 leu 1–32 ura4-D18 otr 1R (dg-glu) Sph::ade6 vgl Δ::KanMX | This study |
| 7. | FY 1180 | h + ade6–210 leu 1–32 ura4-D18 otr 1R (dg-glu) Sph::ade6 | 39 |
| 8. | AB138 | h + Nmyc-clr4 ade6–210 leu1–32 ura4D18 otr1RSph::ade6 Vgl1::HA KanMX | This study |
| 9. | FY 8216 | h + myc3::ago1 leu1–32 ade6–704 ura4-D18 | 39 |
| 10. | AB140 | h + myc3::ago1 leu1–32 ade6–704 ura4-D18 vgl Δ::KanMX | This study |
| 11. | FY 8635 | h + Nmyc-clr4 ade6–210 leu1–32 ura4D18 otr1RSph::ade6 | 39 |
| 12. | FY A2406 | h + rdp1-HA --PFAMX6KAN ade6–210 otr (Sph1):ade6 + | 39 |
| 13. | SPM1079 | h90ade6-M216 leu1–32 ura4-D18 clr4::5XFLAG-clr4 | 39 |
| 14. | AB139 | h90 vgl1 Δ::KanRade6-M216 leu1–32 ura4-D18 clr4::5XFLAG-clr4 | This study |
| 15. | BG-H4839 | vgl1 Δ::KanMX4 ade-M216 ura4-D18 leu1–32 | Bioneer |
| 16. | FY8539 | h + otr1R(SphI):ura4 + ura4-DS/E leu1–32 ade6-M210 dcr1Δ::KanMX | 39 |
| 17. | AB140 | h + Vgl1-HA-HygR otr1R(SphI):ura4 + ura4-DS/E leu1–32 ade6-M210 dcr1Δ::KanMX | This study |
Quantitative RT-PCR
Yeast cells were grown in YES medium (yeast extract supplemented with adenine) at a temperature of 32 °C up to an A600 of 0.5. Total RNA was isolated using the hot phenol method as described previously (2). RNA samples were cleaned to remove potential DNA contaminations using the RNeasy clean-up kit (Qiagen). cDNA was prepared using gene-specific primers for ura4, dg, dh, ade, and actin by using the first-strand cDNA synthesis kit (Thermo Fisher Scientific) according to manufacturer's protocol followed by quantitative PCR using light cycler. Relative RNA levels were calculated from CT values according to ΔCT (Applied Biosystems) after normalization to actin RNA levels.
ChIP assay
ChIP assays were performed as described previously (41, 56). Briefly, 100 ml of cells were grown to an A600 of 1.0. Cells were cross-linked with 10 mm dimethyl adipimidate and subsequently with 1% formaldehyde for 20 min with gentle shaking at room temperature followed by quenching using 125 mm glycine for 5 min. Cell lysis was performed using a bead-beating method. Cross-linked chromatin was sonicated to yield DNA fragments of an average size of 200–600 bp. Immunoprecipitations were carried out using different antibodies (anti-RNA pol II (Abcam); anti-H3K9me2 (Abcam); anti-Swi6 (Abcam); anti-HA (Abcam); anti-FLAG M2 (Sigma); and anti-Myc (Abcam) antibodies). Primers used in the PCR were first analyzed for both efficiency and linearity range by real-time PCR with a LightCycler (Applied Biosystems). All the ChIP experiments were performed three times with similar results. qPCR was performed using SYBR Green reagent (Thermo Fisher Scientific) according to the manufacturer's protocol using a Real Time Cycler (Applied Biosystems). The results shown are based on three independent experiments with standard errors. Data were corrected for the nucleosome occupancy using the total H3 signal.
Silencing assays
Silencing assays were performed as described previously (41). Cells were grown overnight in 5 ml of YEA (yeast extract supplemented with adenine), and 10-fold serial dilutions were prepared for each strain in such a way that the highest density spots contained 1.2 × 105 cells. All the cell dilutions were spotted on normal YEA media plates and TBZ (normal media containing 10 μg/ml TBZ) and FOA plates (normal media containing 1 mg/ml 5-FOA). For otr1::ade6+ silencing assay, cells were spotted on Low Ade plates (YES lacking adenine) for 3 days at 32 °C.
Immunoprecipitations
For immunoprecipitations, nontagged Myc Clr4 and HA Vgl1 cells were grown to an A600 of 2.0, harvested, and resuspended in lysis buffer containing 20 mm Hepes-NaOH, pH 7.5, 100 mm NaCl, 5 mm MgCl2, 1 mm EDTA, pH 8.0, 0.5 mm DTT, 10% glycerol, 0.25% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, complete Protease Inhibitor Mixture. Cells lysis was performed using the bead-beating method. Protein concentrations were normalized using the Bio-Rad protein assay, and the supernatant was incubated with IgG-conjugated Dynabeads for 2 h at 4 °C. Immunoprecipitated material was washed four times with lysis buffer, resuspended in SDS sample buffer, and analyzed by SDS-PAGE followed by Western blotting using anti-HA (Abcam) and anti-Myc 9E10 (Covance) antibodies.
Model generation and validation
The sequences of Clr4 and Vgl1 (S. pombe) were retrieved from UniProt bearing unique and stable entry identifiers O60016 and O59810 respectively. The models were generated from these sequences using the I-TASSER (Iterative Threading ASSEmbly Refinement) server, based on the futuristic algorithms to generate infallible models (57, 58). The Clr4 model was refined by GalaxyRefine, and for Vgl1 the ModRefiner served the purpose. GalaxyRefine used a sequential approach, including side-chain rebuilding and repacking, and finally structure relaxation for improving the global and local structural quality of the models (59). ModRefiner, the algorithm for high-resolution structure refinement, revamps markedly the physical quality of local structures and takes the models close to their native state (60). The stereochemical quality of refined models was checked by generating their respective Ramachandran plots using the accepted PROCHECK of SAVES server (61). Although Clr4 portrayed 88.9% residues in the favored region and 9% residues in the allowed region, Vgl1 showed 78.1% residues and 16.6% residues in these regions, respectively (Fig. 6). These values signify that the models are of good quality and are quite acceptable for in silico studies, including protein–protein docking.
Protein–protein docking
Clr4 was docked against the Vgl1 using the fully-automated web-based protein–protein docking program ClusPro (62, 63). Vgl1 being bigger in size was used as a receptor, whereas Clr4 due to its relatively smaller size was selected as a ligand. The docking was performed using the default parameters, and top 10 docked complexes were generated from which the top model was selected for downstream analysis.
Interaction profile of Vgl1 and Clr4
The docked complex of Vgl1 and Clr4 was uploaded in PDBsum, a webserver with multiple functions providing structural information regarding protein–ligand and protein–DNA contacts. Moreover, this server serves the purpose of finding different interactions between amino acids of two protein chains at the interface (64, 65). The ribbon diagram of Vgl1 and Clr4 was generated using the UCSF Chimera (66).
ChIP with an RNase step
ChIPs with RNase step were performed with slight modifications. Briefly, cells were cross-linked with formaldehyde for 5 min. Cross-linked chromatin was sonicated to yield DNA fragments with an average size of 200–600 bp and treated with treated with 7.5 units of RNase A and 300 units of RNase T1 (RNase A/T1 mixture; Ambion) or an equivalent volume of RNase storage buffer (10 mm Hepes, pH 7.2, 20 mm NaCl, 0.1% Triton X-100, 1 mm EDTA, 50% v/v glycerol). After incubation at 25 °C for 30 min, immunoprecipitations were carried out with a mAb against HA (12CA5).
GST pulldown assay
GST–Clr4 coupled to GSH beads was incubated with recombinant His–Vgl1 (0.5 μg) in 25 mm Hepes (pH 7.5), 100 mm NaCl, 10% glycerol, 100 μg/ml BSA, 0.5 mm DTT, 0.1% Tween 20, and 1 μg/ml leupeptin, pepstatin, and aprotinin overnight at 4 °C on a rotary shaker. Beads were then washed three times with binding buffer, and bound proteins were analyzed by Western blotting using anti-His and anti-GST antibodies.
Author contributions
Z. F. and M. A. designed research; Z. F., E. A., S. B., S. A. G., R. R., and A. M. performed experiments; Z. F. and M. A. analysed the data and wrote the paper.
Acknowledgments
We are grateful to R. Allshire, Jun-ichi Nakayama, Yota Murakami, Danesh Moazed, and Aarti Sevilimedu for yeast strains and plasmids. We thank Dr. Ajazul Hamid Wani for valuable suggestions.
This work was supported by Department of Biotechnology, Government of India, Grants BT/PR16122/BRB/10/1478/2016 and BT/PR22348/BRB/10/1625/2017 and DST-SERB Grant EMR/2016/000958, Government of India (to M. A.); from DST-SERB in the form of Start-Up Grant for Young Scientists YSS/2015/001267 (to S. A. G.); University Grants Commission 23/12/2012(ii) EU-V for Senior Research fellowship (to Z. F.); and DST INSPIRE Grant IF150792 (to R. R.) for doctoral fellowship. The authors declare that they have no conflicts of interest with the contents of this article.
- imr
- innermost repeat
- otr
- outer
- qRT-PCR
- quantitative RT-PCR
- pol
- polymerase
- TBZ
- thiabendazole
- KH
- K homology
- otr
- outer repeat element
- FOA
- 5-fluoroorotic acid
- ChIP
- chromatin immunoprecipitation.
References
- 1. Gonzalez-Sandoval A., and Gasser S. M. (2016) On TADs and LADs, spatial control over gene expression. Trends Genet. 32, 485–495 10.1016/j.tig.2016.05.004 [DOI] [PubMed] [Google Scholar]
- 2. Li B., Carey M., and Workman J. L. (2007) The role of chromatin during transcription. Cell 128, 707–719 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- 3. Tessarz P., and Kouzarides T. (2014) Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 15, 703–708 10.1038/nrm3890 [DOI] [PubMed] [Google Scholar]
- 4. Altaf M., Saksouk N., and Côté J. (2007) Histone modifications in response to DNA damage. Mutat. Res. 618, 81–90 10.1016/j.mrfmmm.2006.09.009 [DOI] [PubMed] [Google Scholar]
- 5. Farooq Z., Banday S., Pandita T. K., and Altaf M. (2016) The many faces of histone H3K79 methylation. Mutat Res Rev Mutat Res. 768, 46–52 10.1016/j.mrrev.2016.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Towbin B. D., Gonzalez-Sandoval A., and Gasser S. M. (2013) Mechanisms of heterochromatin subnuclear localization. Trends Biochem. Sci. 38, 356–363 10.1016/j.tibs.2013.04.004 [DOI] [PubMed] [Google Scholar]
- 7. Cabianca D. S., and Gasser S. M. (2016) Spatial segregation of heterochromatin, uncovering functionality in a multicellular organism. Nucleus. 7, 301–307 10.1080/19491034.2016.1187354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harr J. C., Gonzalez-Sandoval A., and Gasser S. M. (2016) Histones and histone modifications in perinuclear chromatin anchoring from yeast to man. EMBO Rep. 17, 139–155 10.15252/embr.201541809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mekhail K., and Moazed D. (2010) The nuclear envelope in genome organization, expression and stability. Nat. Rev. Mol. Cell Biol. 5, 317–328 10.1038/nrm1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bonasio R., and Shiekhattar R. (2014) Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 48, 433–455 10.1146/annurev-genet-120213-092323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holoch D., and Moazed D. (2015) RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16, 71–84 10.1038/nrg3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Castel S. E., and Martienssen R. A. (2013) RNA interference (RNAi) in the nucleus roles for small RNA in transcription, epigenetics and beyond. Nat. Rev. Genet. 14, 100–112 10.1038/nrg3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ghildiyal M., and Zamore P. D. (2009) Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10, 94–108 10.1038/nrg2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allshire R. C., and Ekwall K. (2015) Epigenetic regulation of chromatin states in Schizosaccharomyces pombe. Cold Spring Harb. Perspect. Biol. 7, a018770 10.1101/cshperspect.a018770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bloom K. S. (2014) Centromeric heterochromatin the primordial segregation machine. Annu. Rev. Genet. 48, 457–484 10.1146/annurev-genet-120213-092033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mizuguchi T., Barrowman J., and Grewal S. I. (2015) Chromosome domain architecture and dynamic organization of the fission yeast genome. FEBS Lett. 7, 2975–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grewal S. I., and Jia S. (2007) Heterochromatin revisited. Nat. Rev. Genet. 8, 35–46 10.1038/nrg2008 [DOI] [PubMed] [Google Scholar]
- 18. Martienssen R., and Moazed D. (2015) RNAi and heterochromatin assembly. Cold Spring Harb. Perspect. Biol. 7, a019323 10.1101/cshperspect.a019323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moazed D. (2009) Small RNAs in transcriptional gene silencing and genome defence. Nature 457, 413–420 10.1038/nature07756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ekwall K., Nimmo E. R., Javerzat J. P., Borgstrøm B., Egel R., Cranston G., and Allshire R. (1996) Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo-domain protein Swi6p and impair centromere function. J. Cell Sci. 109, 2637–2648 [DOI] [PubMed] [Google Scholar]
- 21. Sugiyama T., Cam H., Verdel A., Moazed D., and Grewal S. I. (2005) RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc. Natl. Acad. Sci. U.S.A. 102, 152–157 10.1073/pnas.0407641102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cam H. P., Sugiyama T., Chen E. S., Chen X., FitzGerald P. C., and Grewal S. I. (2005) Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37, 809–819 10.1038/ng1602 [DOI] [PubMed] [Google Scholar]
- 23. Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S. I., and Moazed D. (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303, 672–676 10.1126/science.1093686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Volpe T., and Martienssen R. A. (2011) RNA interference and heterochromatin assembly. Cold Spring Harb. Perspect. Biol. 3, a003731 10.1101/cshperspect.a003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bühler M., Verdel A., and Moazed D. (2006) Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 125, 873–886 10.1016/j.cell.2006.04.025 [DOI] [PubMed] [Google Scholar]
- 26. Buker S. M., Iida T., Bühler M., Villén J., Gygi S. P., Nakayama J., and Moazed D. (2007) Two different argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat Struct Mol Biol. 14, 200–207 10.1038/nsmb1211 [DOI] [PubMed] [Google Scholar]
- 27. Hall I. M., Shankaranarayana G. D., Noma K., Ayoub N., Cohen A., and Grewal S. I. (2002) Establishment and maintenance of a heterochromatin domain. Science 297, 2232–2237 10.1126/science.1076466 [DOI] [PubMed] [Google Scholar]
- 28. Gerace E. L., Halic M., and Moazed D. (2010) The methyltransferase activity of Clr4Suv39h triggers RNAi independently of histone H3K9 methylation. Mol Cell. 39, 360–372 10.1016/j.molcel.2010.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rea S., Eisenhaber F., O'Carroll D., Strahl B. D., Sun Z. W., Schmid M., Opravil S., Mechtler K., Ponting C. P., Allis C. D., and Jenuwein T. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 10.1038/35020506 [DOI] [PubMed] [Google Scholar]
- 30. Nakayama J., Rice J. C., Strahl B. D., Allis C. D., and Grewal S. I. (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 10.1126/science.1060118 [DOI] [PubMed] [Google Scholar]
- 31. Ekwall K., Javerzat J. P., Lorentz A., Schmidt H., Cranston G., and Allshire R. (1995) The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 269, 1429–1431 10.1126/science.7660126 [DOI] [PubMed] [Google Scholar]
- 32. Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., Allshire R. C., and Kouzarides T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo-domain. Nature. 410, 120–124 10.1038/35065138 [DOI] [PubMed] [Google Scholar]
- 33. Iida T., Nakayama J., and Moazed D. (2008) siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol. Cell. 31, 178–189 10.1016/j.molcel.2008.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sadaie M., Iida T., Urano T., and Nakayama J. (2004) A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 23, 3825–3835 10.1038/sj.emboj.7600401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang K., Mosch K., Fischle W., and Grewal S. I. (2008) Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 15, 381–388 10.1038/nsmb.1406 [DOI] [PubMed] [Google Scholar]
- 36. Volpe T. A., Kidner C., Hall I. M., Teng G., Grewal S. I., and Martienssen R. A. (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 10.1126/science.1074973 [DOI] [PubMed] [Google Scholar]
- 37. Motamedi M. R., Verdel A., Colmenares S. U., Gerber S. A., Gygi S. P., and Moazed D. (2004) Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 119, 789–802 10.1016/j.cell.2004.11.034 [DOI] [PubMed] [Google Scholar]
- 38. Jia S., Noma K., and Grewal S. I. (2004) RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304, 1971–1976 10.1126/science.1099035 [DOI] [PubMed] [Google Scholar]
- 39. Bayne E. H., White S. A., Kagansky A., Bijos D. A., Sanchez-Pulido L., Hoe K. L., Kim D. U., Park H. O., Ponting C. P., Rappsilber J., and Allshire R. C. (2010) Stc1: A critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 140, 666–677 10.1016/j.cell.2010.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barrales R. R., Forn M., Georgescu P. R., Sarkadi Z., and Braun S. (2016) Control of heterochromatin localization and silencing by the nuclear membrane protein Lem2. Genes Dev. 30, 133–148 10.1101/gad.271288.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Banday S., Farooq Z., Rashid R., Abdullah E., and Altaf M. (2016) Role of inner nuclear membrane protein complex Lem2–Nur1 in heterochromatic gene silencing. J. Biol. Chem. 291, 20021–20029 10.1074/jbc.M116.743211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burd C. G., and Dreyfuss G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science 265, 615–621 10.1126/science.8036511 [DOI] [PubMed] [Google Scholar]
- 43. Dodson R. E., and Shapiro D. J. (1997) Vigilin, a ubiquitous protein with 14K homology domains, is the estrogen-inducible vitellogenin mRNA 3′-untranslated region-binding protein. J. Biol. Chem. 272, 12249–12252 10.1074/jbc.272.19.12249 [DOI] [PubMed] [Google Scholar]
- 44. Dejgaard K., and Leffers H. (1996) Characterisation of the nucleic-acid-binding activity of KH domains. Different properties of different domains. Eur. J. Biochem. 241, 425–431 10.1111/j.1432-1033.1996.00425.x [DOI] [PubMed] [Google Scholar]
- 45. Cortés A., Huertas D., Fanti L., Pimpinelli S., Marsellach F. X., Piña B., and Azorín F. (1999) DDP1, a single-stranded nucleic acid-binding protein of Drosophila, associates with pericentric heterochromatin and is functionally homologous to the yeast Scp160p, which is involved in the control of cell ploidy. EMBO J. 18, 3820–3833 10.1093/emboj/18.13.3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cortés A., and Azorín F. (2000) DDP1, a heterochromatin-associated multi-KH domain protein of Drosophila melanogaster, interacts specifically with centromeric satellite DNA sequences. Mol. Cell. Biol. 20, 3860–3869 10.1128/MCB.20.11.3860-3869.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huertas D., Cortés A., Casanova J., and Azorín F. (2004) Drosophila DDP1, a Multi-KH-domain protein, contributes to centromeric silencing and chromosome segregation. Curr. Biol. 14, 1611–1620 10.1016/j.cub.2004.09.024 [DOI] [PubMed] [Google Scholar]
- 48. Zhou J., Wang Q., Chen L. L., and Carmichael G. G. (2008) On the mechanism of induction of heterochromatin by the RNA-binding protein vigilin. RNA 14, 1773–1781 10.1261/rna.1036308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weber V., Wernitznig A., Hager G., Harata M., Frank P., and Wintersberger U. (1997) Purification and nucleic-acid-binding properties of a Saccharomyces cerevisiae protein involved in the control of ploidy. Eur. J. Biochem. 249, 309–317 10.1111/j.1432-1033.1997.00309.x [DOI] [PubMed] [Google Scholar]
- 50. Wintersberger U., Kühne C., and Karwan A. (1995) Scp160p, a new yeast protein associated with the nuclear membrane and the endoplasmic reticulum, is necessary for maintenance of exact ploidy. Yeast 11, 929–944 10.1002/yea.320111004 [DOI] [PubMed] [Google Scholar]
- 51. Hirschmann W. D., Westendorf H., Mayer A., Cannarozzi G., Cramer P., and Jansen R. P. (2014) Scp160p is required for translational efficiency of codon-optimized mRNAs in yeast. Nucleic Acids Res. 42, 4043–4055 10.1093/nar/gkt1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bernard P., Maure J. F., Partridge J. F., Genier S., Javerzat J. P., and Allshire R. C. (2001) Requirement of heterochromatin for cohesion at centromeres. Science 294, 2539–2542 10.1126/science.1064027 [DOI] [PubMed] [Google Scholar]
- 53. Allshire R. C., Nimmo E. R., Ekwall K., Javerzat J. P., and Cranston G. (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9, 218–233 10.1101/gad.9.2.218 [DOI] [PubMed] [Google Scholar]
- 54. Volpe T., Schramke V., Hamilton G. L., White S. A., Teng G., Martienssen R. A., and Allshire R. C. (2003) RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 11, 137–146 10.1023/A:1022815931524 [DOI] [PubMed] [Google Scholar]
- 55. Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A. 3rd, Steever A. B., Wach A., Philippsen P., and Pringle J. R. (1998) Heterologous modules for efficient and versatile PCR based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- 56. Altaf M., Utley R. T., Lacoste N., Tan S., Briggs S. D., and Côté J. (2007) Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell. 28, 1002–1014 10.1016/j.molcel.2007.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 1471–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang J., Yan R., Roy A., Xu D., Poisson J., and Zhang Y. (2015) The I-TASSER: Suite protein structure and function prediction. Nat. Methods 12, 7–8 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Heo L., Park H., and Seok C. (2013) GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 41, 3 10.1093/nar/gkt458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xu D., and Zhang Y. (2011) Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys. J. 101, 2525–2534 10.1016/j.bpj.2011.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lovell S. C., Davis I. W., Arendall W. B. 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., and Richardson D. C. (2003) Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins 50, 437–450 10.1002/prot.10286 [DOI] [PubMed] [Google Scholar]
- 62. Kozakov D., Hall D. R., Xia B., Porter K. A., Padhorny D., Yueh C., Beglov D., and Vajda S. (2017) The ClusPro web server for protein–protein docking. Nat. Protoc. 12, 255–278 10.1038/nprot.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Comeau S. R., Gatchell D. W., Vajda S., and Camacho C. J. (2004) ClusPro: a fully automated algorithm for protein–protein docking. Nucleic Acids Res. 32, W96–W99 10.1093/nar/gkh354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Laskowski R. A., Jabłońska J., Pravda L., Vařeková R. S., and Thornton J. M. (2018) Structural summaries of PDB entries. Protein Sci. 27, 129–134 10.1002/pro.3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McDonald I. K., and Thornton J. M. (1994) Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238, 777–793 10.1006/jmbi.1994.1334 [DOI] [PubMed] [Google Scholar]
- 66. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]