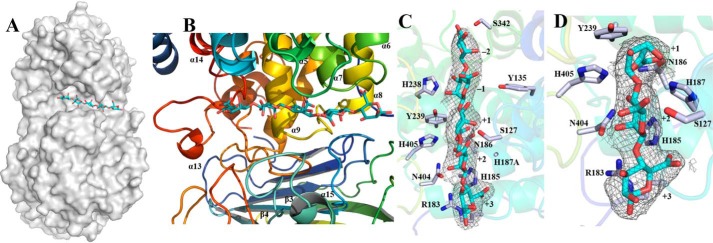
Figure 5.
Substrate-binding groove of TM5. A, surface representation to show the binding site of M5 (shown as sticks and drawn in atom colors (oxygen, red; carbon, cyan)) in the catalytic domain of Dp0100. B, schematic diagrams to show the binding site of M5 and the surrounding elements of secondary structure (colored as in Fig. 3). C and D, electron density surrounding the substrates (shown as in A) in the (2Fo − Fc) omit map (contoured at 1.0σ) in complexes of the H187A mutant with M5 and the WT enzyme with ΔMM, respectively. The sugar-binding subsites are numbered in C and D. Key residues interacting with the substrates are highlighted (drawn in atom colors (oxygen, red; carbon, gray; and nitrogen, blue)) with the remainder of the enzyme shown transparently in cartoon format. Figure was prepared using PyMOL (56).