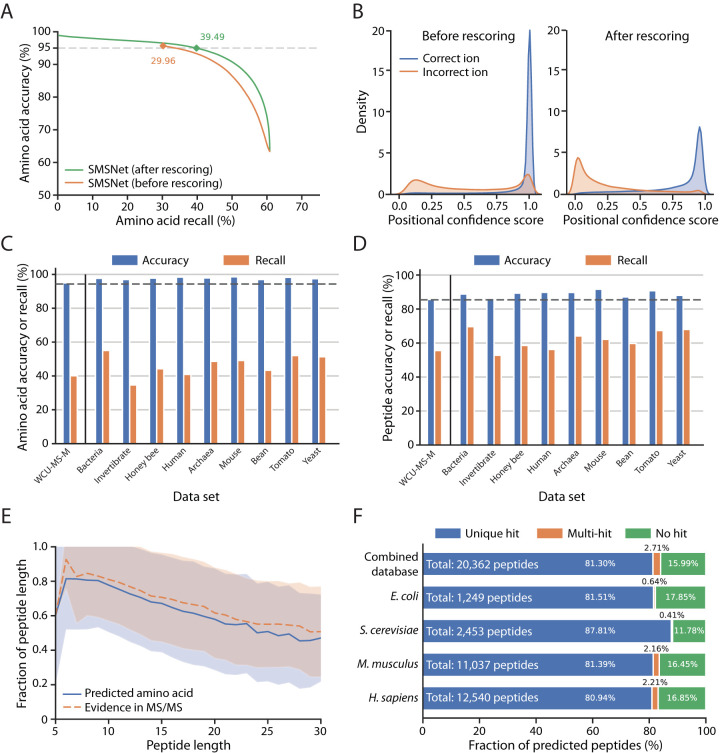
Fig. 3.
The Sequence-Mask-Search framework significantly improves de novo peptide sequencing accuracy. The WCU-MS-M dataset was used to train and evaluate the performance of SMSNet here. A, Amino acid-level performances for SMSNet before and after the positional confidence score adjustment step. The corresponding recalls at 5% amino acid false discovery rate are indicated. B, Histograms showing the distributions of positional confidence scores produced by SMSNet before and after score adjustment. C, Bar plots showing amino acid-level accuracies and recalls of SMSNet on a test set derived from WCU-MS-M and on MS/MS spectra of nine species that comprise the dataset curated by DeepNovo's authors. The threshold on positional confidence score was selected so that 5% amino acid false discovery rate was achieved on the WCU-MS-M test set (the leftmost bars). Dashed line indicates the expected 95% accuracy based on the applied score threshold. D, Similar bar plots showing the results at peptide-level. Dashed line indicates the expected accuracy level based on the applied score threshold. E, Line plots comparing the fraction of identified amino acid positions that pass the same score threshold used in C–D in peptides of various lengths (blue line) to the fraction of amino acid positions that can be definitely determined based on observed ions in the MS/MS spectra (orange dashed line). Shaded area indicates the ±1 standard deviation range. F, Stacked bar plots showing the fraction of identified peptides that could be matched to various protein sequence databases. Amino acid sequence database for each species was downloaded from UniProt (see Experimental Procedures). Combined database integrates amino acid sequences from all four species considered. In each bar, only identifications whose ground truths exist within the corresponding database were counted. “Unique hit” means that the identified sequence matches to exactly one possibility in the database. “Multi-hit” means that the identified sequence matches to multiple possibilities. “No hit” means the identified sequence does not match to anything in the database.