Fig. 4.
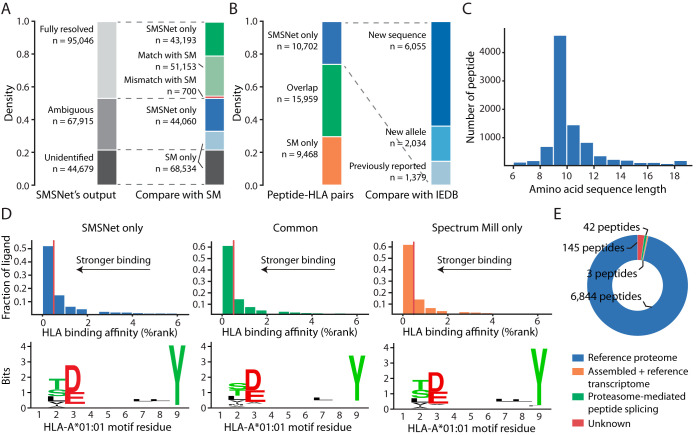
SMSNet uncovers many new HLA antigens. A, Stacked bar plots showing the numbers of MS/MS spectra identified by SMSNet and the overlaps between SMSNet and prior study (17), which utilized Spectrum Mill (SM) software (Agilent Technologies, Inc., Santa Clara, CA) for MS/MS data interpretation. B, Stacked bar plots showing the overlap between peptide-HLA pairs identified by SMSNet and prior study or the Immune Epitope Database (IEDB). “New sequence” indicates that the identified peptide contains amino acid sequence that has not been reported as antigen for any specific HLA allele in IEDB. “New allele” indicates that the identified sequence has been reported to bind to HLA alleles other than the ones considered here. There are 7034 distinct peptides among 10,702 newly identified peptide-HLA pairs. C, The length distribution of 7034 peptides newly identified by SMSNet. D, Histograms and sequence logos comparing the predicted binding affinities and core sequence motifs between peptide-HLA pairs identified by only SMSNet (left), by both SMSNet and prior study (middle), or by prior study only (right). Binding affinities and core motifs were predicted using NetMHCPan. Vertical red lines designate the 0.5% rank threshold typically used to select strong binders. E, Pie chart showing the origins of 7,034 peptides newly identified by SMSNet. Peptide sources were determined by searching amino acid sequences against human proteome, transcriptome, and a database of theoretically possible spliced peptides (see Experimental Procedures).