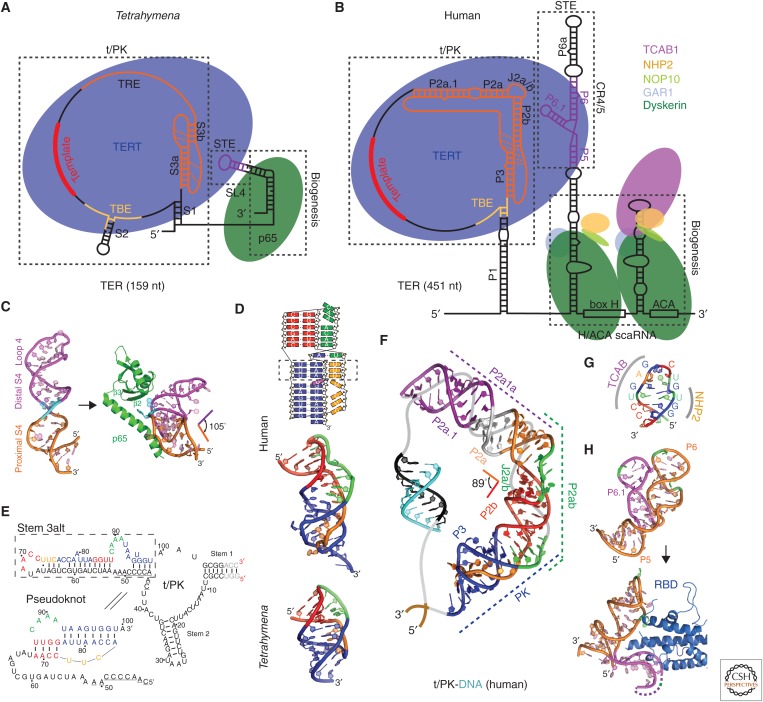
Figure 2.
Tetrahymena and human telomerase core RNP and structures of TER domains. Schematics of telomerase core RNP of (A) Tetrahymena and (B) human telomerase. Proteins are shown at their approximate relative size and interactions, and the shape of TER is based on structural studies described in the text. (C) Nuclear magnetic resonance (NMR) structure of the free Tetrahymena stem-terminus element (STE; stem loop 4) (Protein Data Bank [PDB]: 2FEY) and model of the Tetrahymena p65 atypical RNA recognition motif (xRRM)–stem loop 4 complex based on the crystal structure of p65 xRRM–stem 4 (PDB: 4ERD) and NMR structure of loop 4 (PDB: 2M21). (D) Solution structures of minimal pseudoknots (PKs) from human (PDB: 2K95) and Tetrahymena (PDB: 5KMZ). Secondary structure schematic of the human P2b–P3 minimal PK is shown on the top. Dashed rectangle highlights the three U-A-U triples. (E) Comparison of secondary structures of free Tetrahymena TER and the folded PK, determined by NMR (Cash and Feigon 2017). The template residues are underlined. (F) Molecular model of human template/pseudoknot domain (t/PK). The solution NMR structure of the minimal PK (P2b/P3), P2b–J2a/b–P2a (P2ab), and residual dipolar coupling (RDC)–MC-Sym model of P2a.1–J2a.1–P2a (P2a1a) were computationally combined to model the full-length P2/P3 pseudoknot (Zhang et al. 2010). The single-stranded region (gray) containing the template (black) is shown bound to telomeric DNA (cyan). (G) Solution NMR structure of the 3′ apical stem loop (conserved region 7 [CR7]) containing the TCAB1 and NHP2 binding interfaces, determined by NMR and mutagenesis (Theimer et al. 2007). (H) NMR structure of free medaka (Oryzias latipes) STE (CR4/5) (PDB: 2MHI) compared with crystal structure of the CR4/5–RNA-binding domain (RBD) complex (PDB: 4O26).