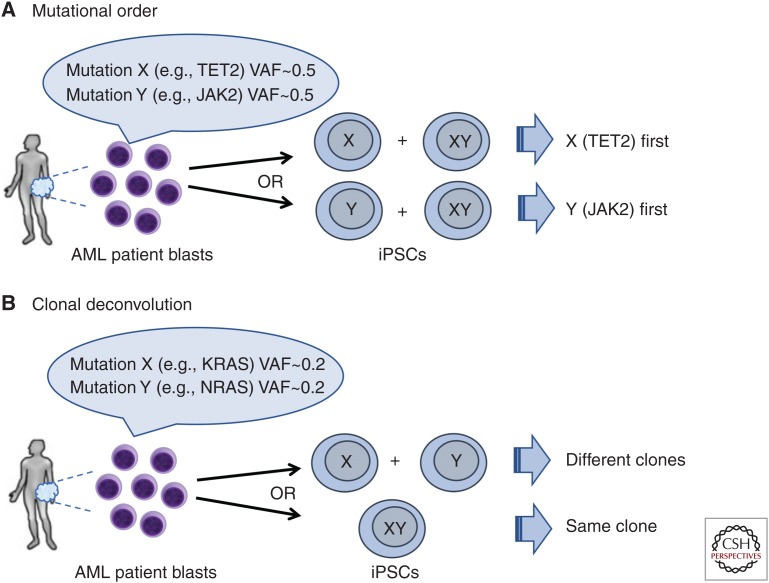
Figure 1.
Mutational order reconstruction and clonal deconvolution by reprogramming. (A) Reprogramming into induced pluripotent stem cells (iPSCs) can decipher the order of mutation acquisition and thus illuminate the clonal history of the disease. In the example shown, two mutations (X and Y, e.g., TET2 and JAK2) have similar allele burden at presentation precluding bulk genetic analyses from determining the order by which they were acquired. iPSCs can capture the precursor single-mutant clone and thus determine which mutation occurred first. (B) Generation of iPSCs can deconvolute the clonal composition of a sample in the case of mutations with low variant allele fractions (VAFs) from the bulk analysis, which cannot determine whether they are present in the same or different clones. In this case, KRAS and NRAS mutations can frequently arise in divergent clonal evolution (example on top) or, alternatively, coexist in the same clone (example in the bottom).