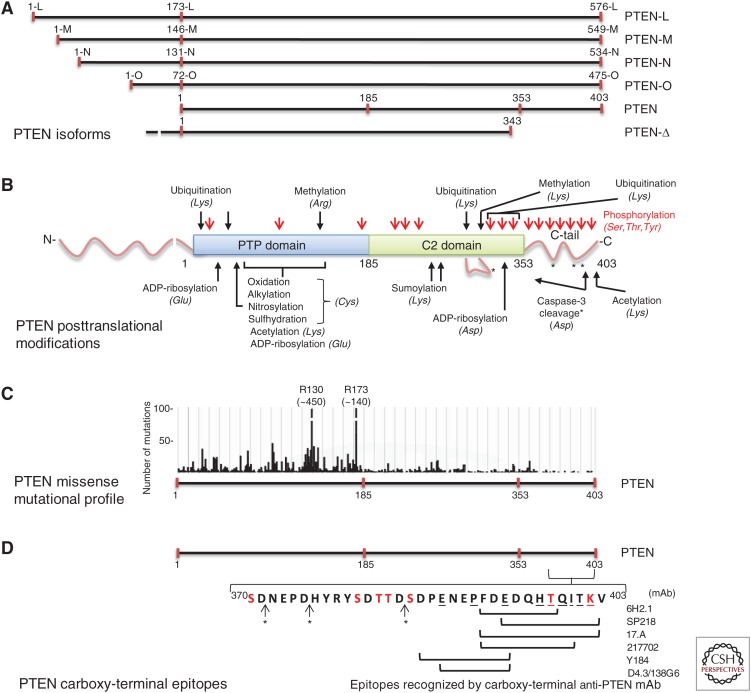
Figure 4.
Variability of PTEN amino acid composition and posttranslational modifications in relation with PTEN carboxy-terminal epitopes. (A) Schematic depiction of PTEN isoforms. PTEN long isoforms (PTEN-L, PTEN-M, PTEN-N, and PTEN-O), generated by alternative translation initiation, are shown in the upper part. Amino acid numbering and nomenclature are according to Pulido et al. (2014) and Tzani et al. (2016). PTEN-Δ isoform (PTEN 1-343-Ser), generated by alternative splicing, is shown in the bottom. PTEN (1–403) is shown in the middle with indication of the residues flanking the PTP and C2 domains and the carboxy-terminal intrinsically disordered region (C-tail). (B) Schematic of PTEN posttranslational modifications. The distinct posttranslational modifications undergone in the different PTEN domains are denoted with indications of the identity of the residues modified as reported for PTEN (1–403; see Table 5 for more information). The disordered region in the C2 domain (residues 286–310) is shown as a line loop. Note that the existence of these modifications in the PTEN long isoforms has not been reported. (C) Number of PTEN missense mutations along PTEN protein found in human tumor samples as annotated in the COSMIC database (Catalogue of Somatic Mutations in Cancer, Wellcome Trust Sanger Institute; Forbes et al. 2017). Note that the y-axis is scaled to allow visualization of low-frequency mutations. The total number of missense mutations for residues R130 and R173 is in parentheses. (D) Delimitation of PTEN carboxy-terminal epitopes recognized by the indicated commercial anti-PTEN mAb. PTEN carboxy-terminal amino acid sequence (residues 370–403) is indicated with a one-letter code. Amino acids in red are subjected to phosphorylation or acetylation (K402). Amino acids underlined are targeted by disease-associated mutations (see Table 6 for more information). *, caspase-3 cleavage sites as shown in B. Epitope mapping is from Mingo et al. (2019) and our unpublished observations.