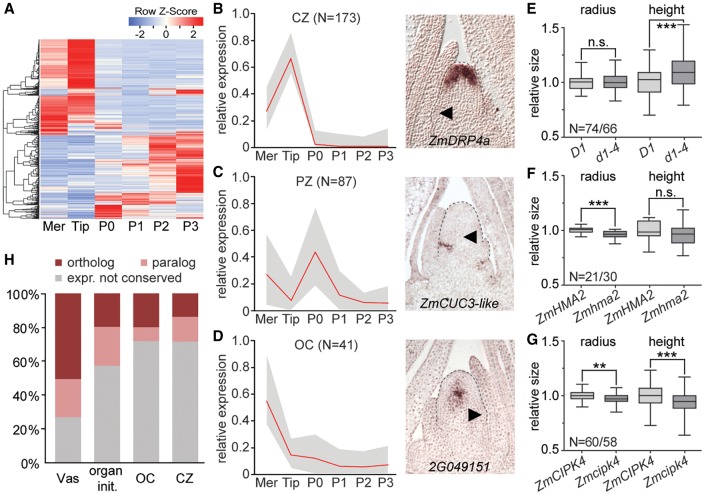
Figure 3.
Divergent gene sets define functional SAM domains in maize versus Arabidopsis. (A) Expression heat map of genes dynamically expressed during the transition from stem cell (Tip) to P3 organ primordium. Genes of Cluster 1 whose expression changes minimally are excluded. (Mer) Meristem. (B–D) Left: composite expression profiles of gene clusters marking the CZ (B), PZ (C), or OC (D). Right: in situ hybridization patterns of select cluster members. Arrowheads, P0; N, number of genes in respective clusters; red line, mean expression; gray profile, range between highest and lowest values. (E–G) Mutations in the OC genes D1 (E), ZmHMA2 (F), and ZmCIPK4 (G) affect meristem height, radius, or both. (N) Number of measured wild-type/mutant individuals. (n.s.) Not significant, (**) P < 0.01, (***) P < 0.001, according to Student's t-test. (H) Percentages of maize genes expressed specifically in vasculature, during organ initiation, or in the OC or CZ with a similarly expressed Arabidopsis ortholog (dark red) or related paralog (light red) (Yadav et al. 2014). Maize genes without an identifiable Arabidopsis ortholog or near paralog are not shown.