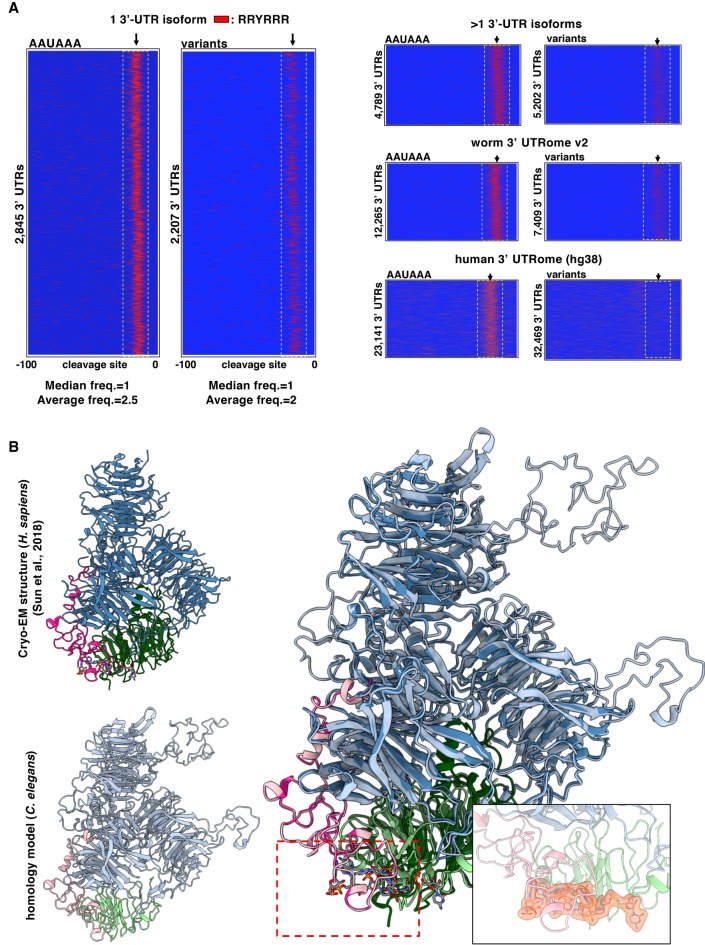
Figure 4.
The sequence requirements of the C. elegans CPSF core complex. (A) PAS element usage of the RRYRRR motif. 3′ UTRs from the 3′ UTRome v2 aligned by their cleavage site in genes with canonical or variant PAS element. Instances of the motif RRYRRR are represented by the thin red bars mapped on 3′-UTR sequences with the coordinates −100 to 0 nt, in which 0 nt represents the cleavage site. The spatial conservation highlighted by the yellow box of this RRYRRR motif is very strong in single 3′-UTR isoforms with canonical PAS elements and is enriched in those with variant PAS elements. This RRYRRR element is maintained in 3′ UTRs that have at least two isoforms, but it is not strongly represented in human 3′-UTR data (hg38) owing to the lack of their annotation. (R) Purine; (Y) pyrimidine. (B) Superimposition of the cryo-EM structure of the previously published human CPSF core complex (Clerici et al. 2018; Sun et al. 2018) to the worm CPSF core complex: CPSF-1 (CPSF1) in blue; PFS-2 (WDR33) in pink; CPSF-4 (CPSF4) in green. The PAS element binding pocket can be fitted into the homology model. The PAS element of the RNA is represented in yellow. The size and the selectivity of the nucleotide-binding pocket can fit other nucleotides as long as the motif is RRYRRR.