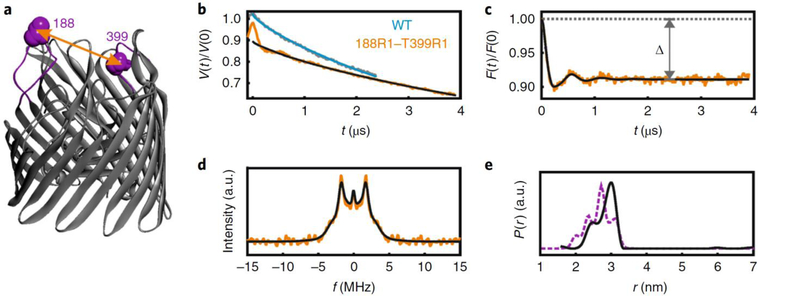
Figure 4 |. In-situ PELDOR in E. coli.
(a) The extracellular loops carrying the positions 188 and 399 are highlighted on the BtuB crystal structure (PDB 1NQH). (b) Original PELDOR data in E. coli as indicated. For WT BtuB (which is naturally Cys-less), the data fit into a stretched exponential decay (), which could not be measured longer due to the weak signal. The data are slightly shifted along the vertical axis for clarity. (c) The dipolar evolution (in yellow) obtained for the 188R1/399R1 PELDOR after correction for the intermolecular contribution () and the corresponding fit from TR (in black). The modulation depth (Δ) value is indicated. Overall, the data suggests a two-dimensional distribution of the spins over the large cell surface and deviation of the value for d (from 2.0) might be for other reasons including the membrane curvature and sample inhomogeneity. (d) The dipolar spectrum obtained with Fourier transformation (in yellow) or TR (in black) of c. (e) Interspin distance distributions obtained from TR of c. The corresponding simulation on the BtuB crystal structure (PDB 1NQH) using the MMM software is overlaid (in violet), which suggests a very good agreement between the conformations observed in the crystal structure and E. coli.