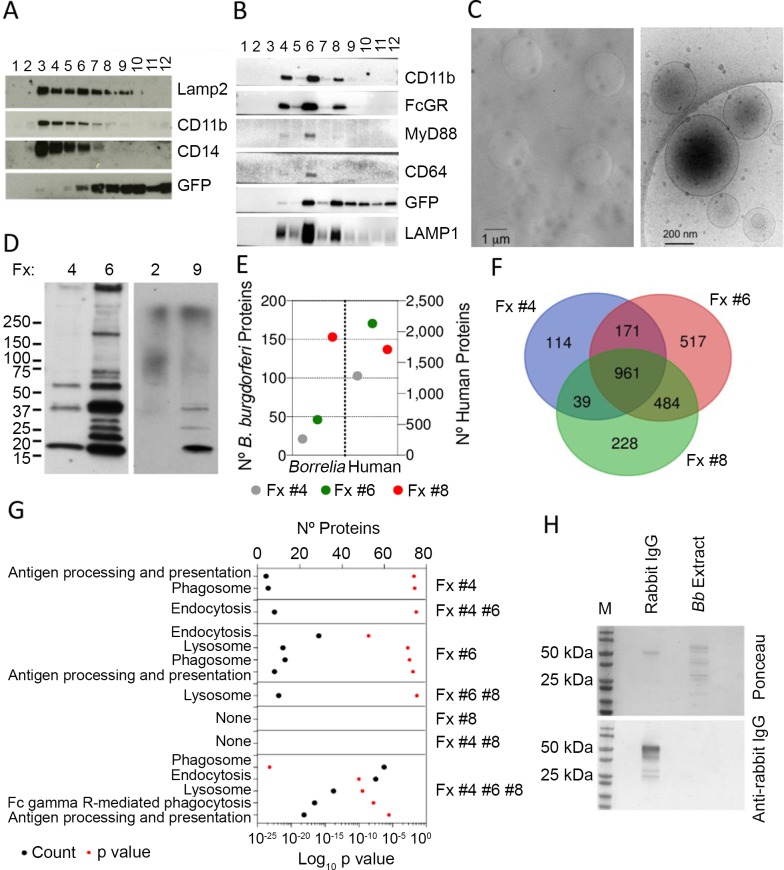
Fig 1. Proteomic characterization of B. burgdorferi-containing, phagosome-enriched fractions.
Immunoblot analysis of B. burgdorferi-containing phagosome enriched sucrose fractions from murine BMMs (A) and human monocyte-derived macrophages (B). The numbers at the top indicate the number of fraction collected from the top. (C) Cryo-electron micrographs of phagosome-enriched fractions from hMACs. The panel on the left shows an image captured at 4,000 X with phagosomes (dark circles). The clear circles represent the holes of the perforated support foil with pre-defined hole size, shape and arrangement. The image on the right shows plasma membrane-derived liposomes at a higher magnification (30,000 X). (D) Immunoblot using B. burgdorferi-infected sera of fractions containing GFP (4, 6, 9) or not from (B). (E) Number of B. burgdorferi and human proteins identified in phagosome-enriched fractions 4, 6 and 8 of hMACs. (F) Venn diagram showing the overlap in protein composition of hMAC phagosome-enriched fractions 4, 6 and 8. A total of 2514 proteins were identified in all 3 fractions, with 961 shared proteins. (G) KEGG pathway analysis of the proteins identified in each fraction. The proteins analyzed correspond to those represented only in each individual fraction, those shared by 2 fractions and the 961 common proteins found in all 3 fractions, as in Fig 1D. (H) Immunoblot of B. burgdorferi extracts (5 μg) obtained from preparations used in the phagocytosis assays. Rabbit IgG (1 μg) was used as control. The blots were tested with an anti-rabbit IgG antibody. The upper panel shows the staining with Ponceau.