Figure 3. The Mus musculus domesticus Bacterial Gut Microbiome can be Transferred to Pregnant GF C57BL/6 Mice and Maintained in their Multigenerational Offspring.
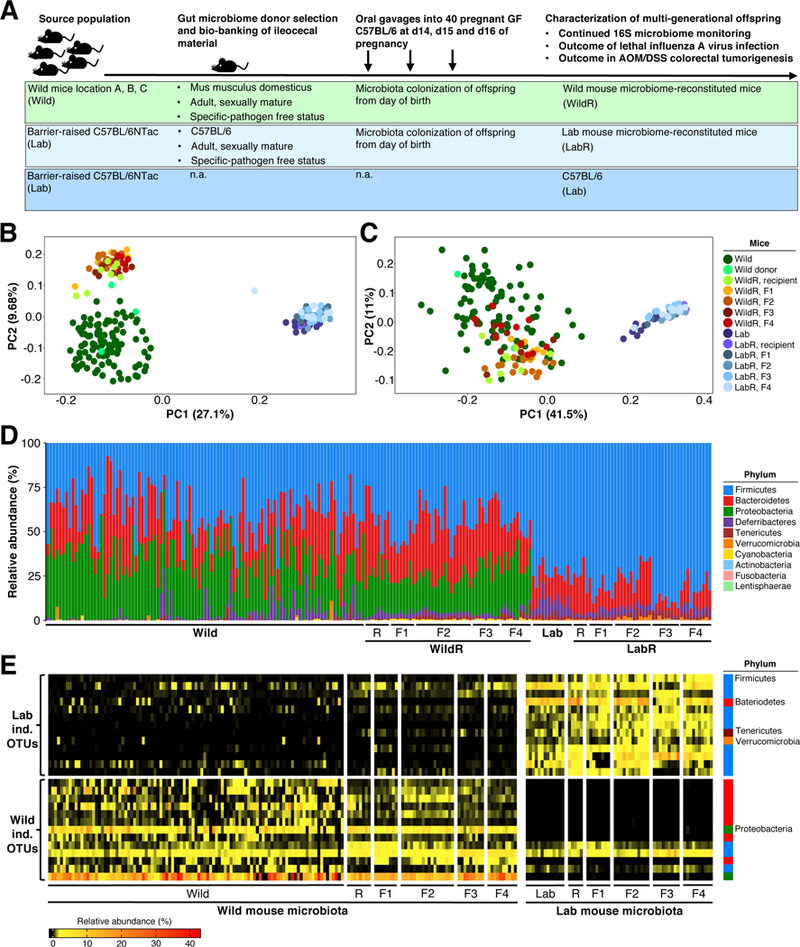
(A) Experimental strategy. (B – E) 16S rRNA gene profiling data comparing the gut microbiome of Mus musculus domesticus (Wild) or C57BL/6NTac (Lab), the recipient mice (WildR and LabR recipients [R]), and their subsequent generations (WildR F1, F2, F3, F4 and LabR F1, F2, F3, F4). (B) Unweighted UniFrac PCoA. (C) Weighted UniFrac PCoA. (D) Relative abundance at the rank of phylum. (E) Indicator species analysis identified bacterial OTUs of the investigated Wild and Lab gut microbiomes. Heatmap shows the relative abundances of Wild indicative and Lab indicative OTUs from all sampled generations.See also Figures S5, Figure S6 and Table S6.
