Abstract
Five bioactive Annonaceous acetogenins, including three new compounds, annonamuricins A (1), B (2), and C (3), one registered but no spectral data reported compounds, annonamuricin D (4), and one known compound annonacin (5) were isolated from Graviola fruit (Annona muricata) and further determined through bioassay-guided fractionation. All five compounds are C35 Anonnonaceous acetogenins with a mono-tetrahydrofuran ring and four hydroxyls. Their structures were elucidated using spectral methods as well as chemical modification after isolation via chromatographic techniques and HPLC purification. These acetogenins demonstrated potent anti-proliferative activities against human prostate cancer PC-3 cells.
Keywords: Annonamuricin, Annona muricata, Human prostate cancer PC-3 cells, Anonnonaceous acetogenins
Introduction
Annonaceous acetogenins are a unique set of derivatives of C35 or C37 long chain fatty acids derived from the polyketide pathway1 and characteristic of family Annonaceae. These phytochemicals have been reported to possess significant antitumor properties. They show selective toxicity to various types of cancer cells, including multi-drug resistant cancer cell lines, at very low dosages.2–9 The most potent acetogenins inhibited the growth of Adriamycin-resistant human mammary adenocarcinoma (MCF-7/Adr) cells with a potency ~250 times over that of Adriamycin.10 Studies on the pharmacological mechanisms underlying the aforementioned functions indicated that Annonaceous acetogenins induced cytotoxicity by inhibiting the mitochondrial complex I of the electron transport chain which is involved in ATP synthesis.5 Mitochondrial complex I could be a potential target in cancer therapeutics due to the higher demand for ATP in tumor cells than normal cells. Hence, these compounds are regarded as promising anti-cancer agents worthy of further animal studies, with attempts of incorporation into new chemotherapeutic drugs.
Graviola (Annona muricata) is a tropical fruit tree of the family Annonaceae. The fruit pulp has sweet flesh and a distinctive flavor that can be eaten out of hand and is ideal for making drinks, candy, sorbets and ice cream. In addition, the fresh fruit and fruit juice are often consumed as folk medicines for cooling fevers, increasing mother’s milk supply after childbirth, and as an astringent for diarrhea and dysentery. The crushed seeds are used against internal and external parasites, head lice, and worms. Additionally, the barks, leaves, and roots of Graviola are all regarded as natural medicine in the indigenous tropics. The barks, leaves, and roots have been used to manage a wide range of human diseases including inflammatory conditions, rheumatism, neuralgia, diabetes, hypertension, insomnia, cystitis, parasitic infections, and cancer.11 At the present time most attention has been focused on Graviola’s anti-cancer properties associated with Annonaceous acetogenins found in its extracts. Specific acetogenins in Graviola have been reported to have selective toxicity in vitro to the following types of tumor cells: lung carcinoma, human breast solid tumor, prostate adenocarcinoma, pancreatic carcinoma, colon adenocarcinoma, liver cancer, human lymphoma and multi-drug resistant human breast adenocarcinoma.1,6,7,12–14
In previous studies of Graviola-derived Annonaceous acetogenins, the stems, leaves, and seeds were found to contain more than 40 acetogenins,15 and more are continuously being discovered. 6,13 Graviola products, namely capsules and tinctures, are becoming more widely available in the U.S. markets, and are now offered under several different manufacturer labels in health food stores. The therapeutic dosage of Graviola leaf is reported to be 2–3 g, taken 3 or 4 times daily.14 We have previously reported that the extract of Graviola fruit has significant activity against selected breast cancer cell lines,16 and the active fractions appear to possess characteristic chemical properties of Annonaceous acetogenins. This study also showed that Graviola fruit extract exerted the selective anti-tumorigenic effect against breast cancer cells by down-regulating the expression of the epidermal growth factor receptor.16 In vitro and in vivo studies reported that Graviola extracts act as inhibitors of multiple signaling pathways that regulate metabolism, cell cycle, survival, and metastatic properties of various prostate cancer cells.14,17 To our knowledge, most of the previously-reported natural compounds were isolated from Graviola’s leaf and stem, bark, and seeds. Little chemical studies on the widely consumed fruit pulp have been conducted. Our studies have also found that there are differences in acetogenins between the provenances of Graviola fruits. On the basis of our previous separation work,18,19 we chose one Graviola fruit pulp preparation with diverse acetogenins, performed bioassay-guided fractionation, and further purified and then identified five more acetogenins (Fig. 1) in this current study. Among them, three new, one registered but no spectral data reported, and one previously discovered Annonaceous acetogenins were determined and exhibited inhibitory activities against human prostate cancer PC-3 cell lines.
Fig. 1.
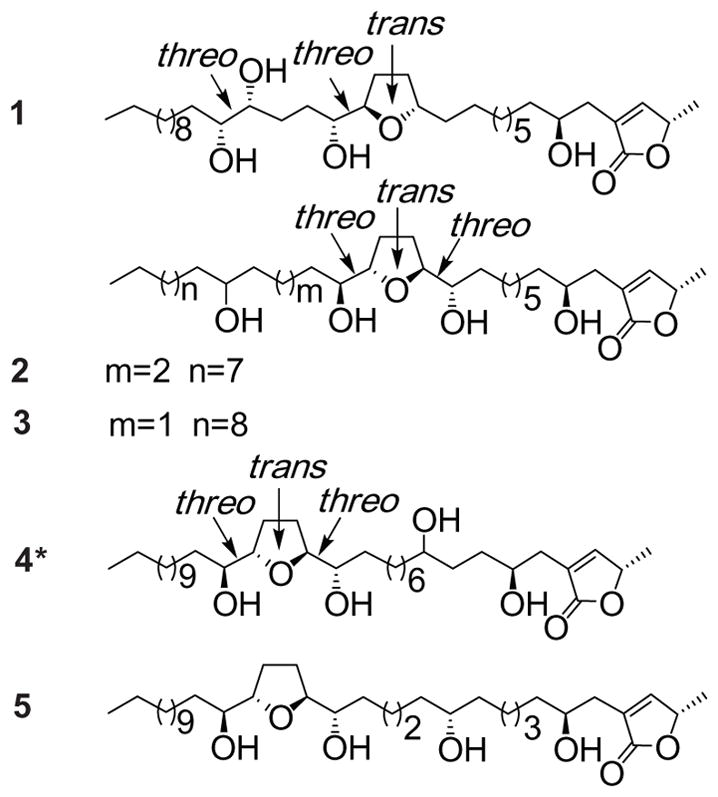
Structures of annonacous acetogenins, annonamuricins A (1), B (2), C (3), D (4) and annonacin (5) from Graviola fruit.
Result and discussion
Three new, one registered, and one known mono-THF acetogenins were isolated and purified from EtOH extract of Graviola fruit powder using silica gel open column and semi-preparative reverse-phase HPLC. Annonamuricin A (1) was isolated and purified from fraction GF2-12, and annonamuricins B (2) and C (3) together with annonamuricin D (4) and annonacin (5)20 were from fraction GF3-19. All pure compounds appear as waxy solids.
By the comparisons of 1H and 13C NMR data of five compounds (1–5) with those reported for known acetogenins, the terminal α,β-unsaturated γ-1actone unit with a 4-OH, typical of many Annonaceous acetogenins, exists in all three compounds (see Table 1).21,22 The four peaks from δ 69 to 75 in each of their 13C NMR spectra suggested the existence of four hydroxyls in each compound.21–23 The 13C NMR signals of the THF oxymethines are usually located between δ 79 and 85,21–23 and a count of peaks in this region revealed all compounds had one THF ring.24,25 The 1H and 13C NMR data of compounds 2 and 3 (Table 1) were almost the same, however, their retention times were obviously different in HPLC. The HRESIMS results gave all three compounds the same quasi-molecular ion, the molecular composition was all C35H64O7, however, their ESIMS gave the different fragmentations which helped to characterize their diverse planar structures. The stereochemistry of hydroxyls in 1–3 was indicative by the 1H chemical shift and tetra-MTPA derivatives.
Table 1.
NMR data (δ values) of compounds 1–4 (400 MHz for 1H;100 MHz for 13C, in CDCl3).
| Position | Compd. 1 | Compd. 2 | Compd. 3 | Compd. 4 | ||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||
| 1H | 13C | 1H | 13C | 1H | 13C | 1H | 13C | |
| 1 | 175.1 | 174.7 | 174.7 | 174.7 | ||||
| 2 | 130.6 | 130.9 | 131.1 | 131.1 | ||||
| 3 | 2.36 dd (15.2, 8.0), 2.50 dd (15.2, 1.6) | 152.3 | 2.35 dd (15.6, 8.0), 2.46 dd (15.6, 2.0) | 152.0 | 2.35 dd (15.6, 8.0), 2.46 (dd, (15.6, 2.0) | 151.9 | 2.40 dd (14.8, 8.0), 2.51 (dd, (14.8, 2.0) | 152.0 |
| 4 | 3.79 m | 69.2 | 3.79 m | 69.5 | 3.79 m | 69.8 | 3.79 m | 69.7 |
| 5 | 1.22–1.71 m | 37.2 | 1.25–1.62 m | 37.2 | 1.30–1.70 m | 37.2 | 1.24–1.66 m | 36.9–37.4 |
| 6 | 1.22–1.71 m | 25.5–25.7 | 1.25–1.62 m | 29.3–29.7 | 1.30–1.70 m | 29.2–29.7 | 1.24–1.66 m | 36.9–37.4 |
| 7 | 1.22–1.71 m | 29.3–29.7 | 1.25–1.62 m | 29.3–29.7 | 1.30–1.70 m | 29.2–29.7 | 3.40 m | 71.4 |
| 8 | 1.22–1.71 m | 29.3–29.7 | 1.25–1.62 m | 29.3–29.7 | 1.30–1.70 m | 29.2–29.7 | 1.24–1.66 m | 36.9–37.4 |
| 9 | 1.22–1.71 m | 29.3–29.7 | 1.25–1.62 m | 29.3–29.7 | 1.30–1.70 m | 29.2–29.7 | 1.24–1.66 m | 25.4–25.6 |
| 10 | 1.22–1.71 m | 29.3–29.7 | 1.25–1.62 m | 25.6 | 1.30–1.70 m | 25.5–25.6 | 1.24–1.66 m | 29.4–29.7 |
| 11 | 1.22–1.71 m | 26.1 | 1.25–1.62 m | 31.8–33.8 | 1.30–1.70 m | 31.9–34.0 | 1.24–1.66 m | 29.4–29.7 |
| 12 | 1.22–1.71 m | 35.6 | 3.40 m | 74.0–74.3 | 3.40 m | 74.1–74.3 | 1.24–1.66 m | 29.4–29.7 |
| 13 | 3.78 q (7.2) | 79.5 | 3.79 m | 82.6–82.7 | 3.79 m | 82.4–82.7 | 1.24–1.66 m | 25.4–25.6 |
| 14 | 1.50–2.02 m | 28.4 | 1.61–1.98 m | 28.0–28.8 | 1.61–1.99 m | 28.0–28.8 | 1.24–1.66 m | 31.9–34.0 |
| 15 | 1.50–2.02 m | 31.9–33.4 | 1.61–1.98 m | 28.0–28.8 | 1.61–1.99 m | 28.0–28.8 | 3.40 m | 74.0–74.2 |
| 16 | 3.79 m | 81.9 | 3.79 m | 82.6–82.7 | 3.79 m | 82.4–82.7 | 3.79 m | 82.6–82.7 |
| 17 | 3.38 m | 74.0–74.3 | 3.40 m | 74.0–74.3 | 3.40 m | 74.1–74.3 | 1.61–1.98 m | 28.2–28.8 |
| 18 | 1.22–1.71 m | 31.9–35.4 | 1.25–1.62 m | 31.8–33.8 | 1.30–1.70 m | 31.9–34.0 | 1.61–1.98 m | 28.2–28.8 |
| 19 | 1.22–1.71 m | 31.9–35.4 | 1.25–1.62 m | 25.6 | 1.30–1.70 m | 25.5–25.6 | 3.79 m | 82.6–82.7 |
| 20 | 3.38 m | 74.0–74.3 | 1.25–1.62 m | 25.6 | 1.30–1.70 m | 37.2 | 3.40 m | 74.0–74.2 |
| 21 | 3.38 m | 74.0–74.3 | 1.25–1.62 m | 37.2 | 3.40 m | 71.4 | 1.24–1.66 m | 31.9–33.4 |
| 22 | 1.22–1.71 m | 31.9–35.4 | 3.40 m | 71.3 | 1.30–1.70 m | 37.2 | 1.24–1.66 m | 25.4–25.6 |
| 23 | 1.22–1.71 m | 25.5–25.7 | 1.25–1.62 m | 37.2 | 1.20–1.60 m | 25.5–25.6 | 1.24–1.66 m | 29.4–29.7 |
| 24 | 1.22–1.71 m | 29.3–29.7 | 1.25–1.62 m | 25.6 | 1.20–1.60 m | 29.3–29.7 | 1.24–1.66 m | 29.4–29.7 |
| 25–29 | 1.22–1.71 m | 29.3–29.7 | 1.25–1.62 m | 29.3–29.7 | 1.20–1.60 m | 29.3–29.7 | 1.24–1.66 m | 29.4–29.7 |
| 30 | 1.22–1.71 m | 31.9 | 1.25–1.62 m | 31.8–33.8 | 1.20–1.60 m | 31.9–34.0 | 1.24–1.66 m | 31.9–33.4 |
| 31 | 1.22–1.71 m | 22.7 | 1.25–1.62 m | 22.6 | 1.20–1.60 m | 22.7 | 1.24–1.66 m | 22.7 |
| 32 | 0.84 t (6.8) | 14.1 | 0.88 t (7.0) | 14.1 | 0.88 t (6.8) | 14.1 | 0.86 t (6.8) | 14.1 |
| 33 | 7.16 s | 152.3 | 7.08 s | 152.0 | 7.17 s | 152.0 | 7.18 s | 152.0 |
| 34 | 5.03 qd (6.4, 1.2) | 78.2 | 5.06 m | 78.0 | 5.02 qd (6.4, 1.2) | 78.0 | 5.05 qd (6.4, 1.2) | 78.1 |
| 35 | 1.41 d (6.4) | 19.1 | 1.42 d (6.4) | 19.0 | 1.40 d (7.2) | 19.1 | 1.40 d (7.2) | 19.1 |
In the 1H and 13C NMR spectra of compound 1, δH 3.79/δC 69.2, in addition to the signals of a methylene at δH 2.36 (dd, J = 14.8, 8.0 Hz, 1H) and 2.50 (dd, J = 14.8, 1.6 Hz, 1H), indicated the presence of a hydroxyl group at C-4. Signals were observed at δH 3.79 (m, 2H)/δC 79.5, 81.9, and δH 3.38 (m, 3H)/δC 74.5, 74.4, 74.1, which were consistent with a THF ring with one flanking hydroxyl group in a threo configuration and a threo 1,2-diol group.26,27 The NMR spectral data were the similar as that squadiolin C,27 however, there was difference between their mass fragmentations.
In the HRESIMS spectrum of compound 1, the peaks at m/z 635.4275 [M+K]+ (cal. 635.4289) and 619.4557 [M+Na]+ (cal. 619.4550) indicated a molecular weight of 596 and molecular formula of C35H64O7. In the EIMS spectrum, the fragments [m/z 394 (393), 353, 323 (322), 186] suggested that the THF ring located between C-13/C-16 and one diol group at C-20/C-21 (see Scheme 1).
Scheme 1.
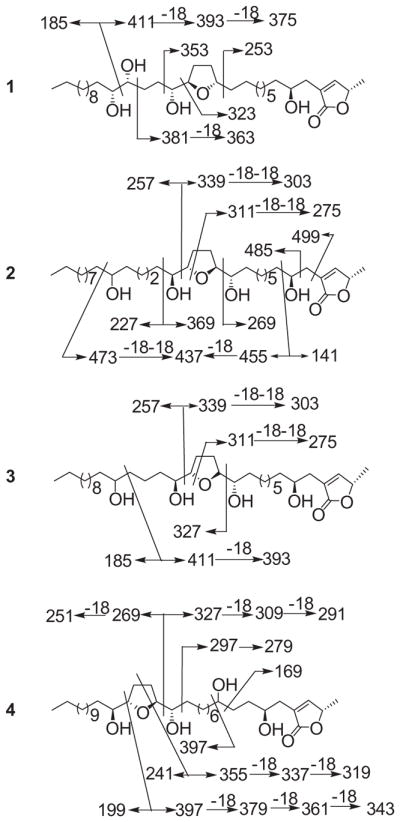
MS fragmentation (m/z values) of annonacous acetogenins from Graviola fruit.
1H NMR data of tetra-Mosher ester derivatives were assigned according to regular, single-relayed and double-relayed COSY spectra (see Table 2). The negative value of δH (S-R) at H-13 (−0.13) suggested an R configuration at C-17. The positive value of H-16 (−0.18) indicated that the configurations at C20/21 were R/R.25 Thus, together with the aforementioned α,β-unsaturated γ-lactone ring, the structure (Fig. 1) of 1 was determined and the compound was named annonamuricin A.
Table 2.
Some selected 1H NMR δ values of tetra-MTPA derivatives of compounds 1–3.
| 1S | 1R | ΔδH(S-R) | 2S | 2R | ΔδH(S-R) | 3S | 3R | ΔδH(S-R) | |
|---|---|---|---|---|---|---|---|---|---|
| 12 | – | – | – | 5.35 | 5.40 | −0.06 | 5.39 | 5.38 | −0.01 |
| 13 | 3.82 | 3.95 | −0.13 | 3.90 | 4.11 | −0.11 | 4.02 | 4.14 | −0.12 |
| 16 | 3.95 | 4.13 | −0.18 | 4.24 | 4.26 | −0.02 | 4.26 | 4.23 | −0.03 |
| 17 | 4.18 | 4.19 | −0.01 | 5.73 | 5.70 | +0.03 | 5.73 | 5.61 | +0.12 |
| 20 | 5.04 | 4.98 | +0.06 | – | – | – | – | – | – |
In the 1H and 13C NMR spectra of compound 2, two sets of signals at δH 3.79 (3H)/δC 82.7, 82.6 and 69.5, δH 3.40 (2H)/δC 74.5 and 74.4, in addition to signals at δH 1.98 (2H) and 1.61 (2H)/δC 28.8, indicated the presence of one THF ring with two flanking hydroxyl groups in a relative threo/trans/threo configuration (Table 1).28 Furthermore, a signal at δH 3.40 (1H)/δC 71.3 indicated the presence of an isolated hydroxyl group in compound 2.26 A [M+Na]+ peak at m/z 619.4550 and a [M+H]+ peak at m/z 597.4727 in the HRESIMS of compound 2 appeared to be consistent with the molecular formula C35H64O7.
Close examination of the ESIMS fragmentation of 2, m/z 339 (338)/258, and 411–393/185, indicated that the THF ring was located C-13/C-16 based on a peak at m/z 269 (C-12/C-13 cleavage) and a peak at m/z 339 (C-16/C-17 cleavage). The hydroxyl group was present at C-22 according to ESIMS peaks at m/z 473 (C-22/C-23 cleavage) and 437 (C-22/C-23 cleavage – H2O) (Scheme 1). Thus, the structure of 2 was fully determined as shown in Fig. 1, and the compound has been given the name annonamuricin B.
In the 1H and 13C NMR spectra of compound 3, two sets of signals at δH 3.78 (3H)/δC 82.7, 82.5 and 69.8, δH 3.39 (2H)/δC 74.3 and 74.1, in addition to signals at δH 1.97 (2H) and 1.66 (2H)/δC 28.8, indicated the presence of one THF ring with two flanking hydroxyl groups in a relative threo/trans/threo configuration (Tables 1 and 2).23,28 In addition, a signal at δH 3.40 (1H)/δC 71.4 indicated the presence of an isolated hydroxyl group in compound 3.26 A [M+H]+ peak at m/z 597.4724 and a [M+Na]+ peak at m/z 619.4445 in the HRFABMS of compound 3 were consistent with the molecular formula C35H64O7.
Close examination of the ESIMS fragmentation of 3, peaks at m/z 257 and 339 (C-16/C-17 cleavage), m/z 327 (C-12/C-13 cleavage), indicated that the THF ring was located between C-13/C-16. Peaks at m/z 393 (C-20/C-21 cleavage – H2O) and 185 (Scheme 1) suggested that the hydroxyl group was at C-21. Thus, the structure of 3 was fully determined as shown in Fig. 1, and the compound was given the name annonamuricin C.
In the 1H and 13C NMR spectra of compound 4, two sets of signals at δH 3.79 (3H)/δC 82.7, 82.6 and 69.7, δH 3.39 (2H)/δC 74.1 and 74.0, in addition to signals at δH 1.97 (2H) and 1.66 (2H)/δC 28.8, indicated the presence of one THF ring with two flanking hydroxyl groups in a relative threo/trans/threo configuration (Tables 1 and 2).23,28 Furthermore, a signal at δH 3.40 (1H)/δC 71.7 indicated the presence of an isolated hydroxyl group in compound 4.26 A [M+H]+ peak at m/z 597.4722 and a [M+Na]+ peak at m/z 619.4545 in the HRFABMS of compound 4 were consistent with the molecular formula C35H64O7.
Close examination of the ESIMS fragmentation of 4, peaks at m/z 327 (C-15/C-16 cleavage and 309 (C-15/C-16 cleavage – H2O), m/z 397 (C-19/C-20 cleavage) and its series of fragments lost H2O, m/z 379, 361, and 343 indicated that the THF ring was located between C-16/C-19. Peaks at m/z 169 (C-6/C-7 cleavage) and 397 (C-7/C-8 cleavage) (Scheme 1) suggested that the hydroxyl group was at C-21. Thus, the structure of 4 was fully determined as shown in Fig. 1, it has been registered as CAS 1026694-89-2, however, there was no name and any reference found, and the compound was given the name annonamuricin D.
All above C35 acetogenins derived from the polyketide pathway, were in agreement with the accepted hypotheses on biosynthesis of mono-THF ring acetogenins acetogenins.29 Annonamuricin A (1) presumably generated through the biogenetic pathway of gigantetrocin A-type acetogenins, a mono-THF ring with one flanking hydroxyls cyclized from a keto cis-alkene through reduction of the keto group, the vicinal diol formed by ring opening of another epoxide group; whereas annonamuricins B–D (2–4) arised from the biogenetic pathway of annonacin-type acetogenins, a mono-THF ring with two flanking hydroxyls cyclized by the ring opening of epoxide moieties from the epoxidation of cis-cis dienes (Scheme 2).
Scheme 2.
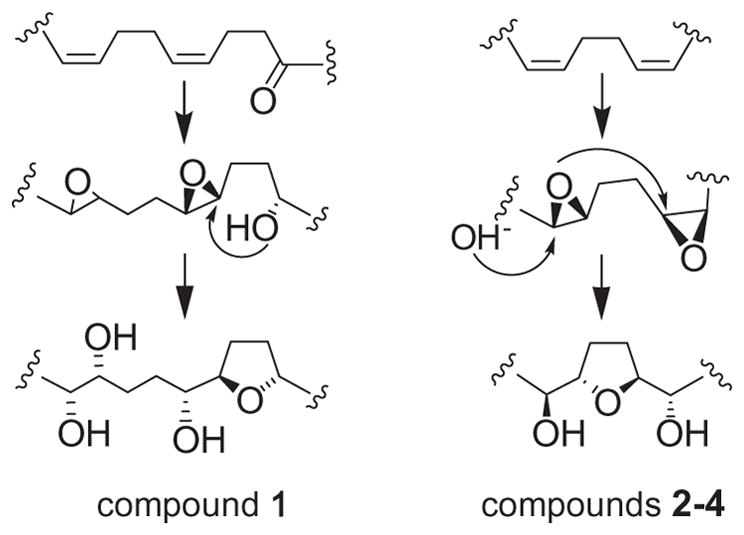
The possible biosynthetic pathways of compounds 1–4.
The newly isolated and elucidated annonamuricins A (1), B (2), C (3), D (4) and annonacin (5) were evaluated for anti-proliferative activity against human prostate cancer PC-3 cells. They all showed inhibitions, with the following potency: annonamuricin C (3) > annonacin (5) > annonamuricin B (2) > annonamuricin D (4) > annonamuricin A (1) (Fig. 2), which indicated that the structure-activity relationship (SAR) of acetogenins was not only affected by the number of hydroxyl groups27 but also by the positions of these groups. It was reported that annonacin was one of the active compounds in Graviola responsible for its anti-proliferative activities against multiple tumor cell lines.30 The compound exhibited strong growth inhibition on PC-3 cells in our experiment, where cell viability was reduced by 96.9% after treatment at 20 μg/ml (Fig. 2). Other three compounds, annonamuricin B, C and D showed comparable anti-proliferative activities, where cell viability was reduced by 96.9, 97.7, and 96.0%, respectively. While the anti-proliferative activity of annonamuricin A was significantly lower than other acetogenins, it still was able to inhibit cell growth by 86.0% at 20 μg/ml.
Fig. 2.
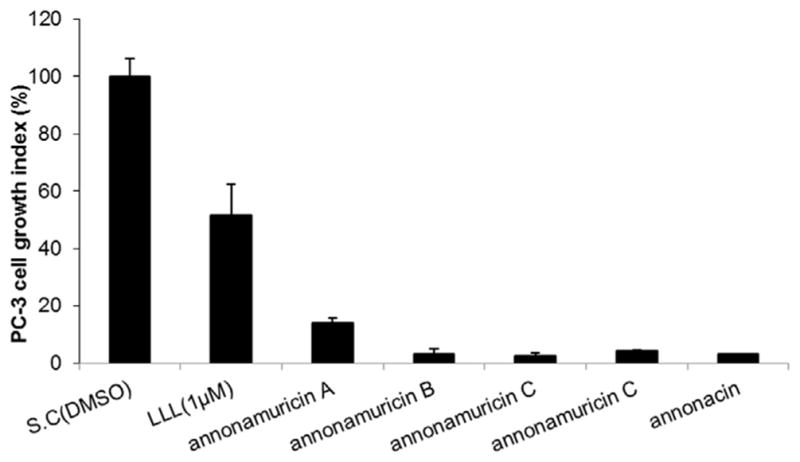
Anti-proliferative activity of graviola-derived annonamuricins A, B, C, D and annonacin against human prostate cancer cells (MTT assay). 60% confluent PC3 cells were incubated with the compounds at 20 μg/ml or positive control (LLnL, N-acetyl-L-leucinyl-L-norleucinal) at 1 μM for 24 h (annonamuricin A by 80% full PC3 cells for 48 h). DMSO served as solvent control. By LSD multiple comparison test in SPSS, the bioactive differences were labeled with A, B, C (at the 0.01 level), and d (at the 0.05 level).
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Center for Complementary and Integrative Health (NCCIH, formerly the National Center for Complementary and Alternative Medicine [NCCAM]) of the National Institutes of Health under Award Number R01AT007566. The authors would like to thank Dr. Brian Shay of Central Instrument Facility, Wayne State University for HR-ESI-MS analysis; and Dr. James Windak and Dr. Paul Lennon of University of Michigan for EI-MS analysis.
A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.tetlet.2017.04.016.
References
- 1.Kim GS, Zeng L, Alali F, et al. Phytochemistry. 1998;49:565. doi: 10.1016/s0031-9422(98)00172-1. [DOI] [PubMed] [Google Scholar]
- 2.Tormo JR, Royo I, Gallardo T, et al. Oncol Res. 2003;14:147. doi: 10.3727/000000003771013099. [DOI] [PubMed] [Google Scholar]
- 3.Tormo JR, DePedro N, Royo I, et al. Oncol Res. 2005;15:129. doi: 10.3727/096504005776367915. [DOI] [PubMed] [Google Scholar]
- 4.Oberlies NH, Jones JL, Corbett TH, Fotopoulos SS, McLaughlin JL. Cancer Lett. 1995;96:55. doi: 10.1016/0304-3835(95)92759-7. [DOI] [PubMed] [Google Scholar]
- 5.McLaughlin JL. J Nat Prod. 2008;71:1311. doi: 10.1021/np800191t. [DOI] [PubMed] [Google Scholar]
- 6.Chang FR, Wu YC. J Nat Prod. 2001;64:925. doi: 10.1021/np010035s. [DOI] [PubMed] [Google Scholar]
- 7.Liaw CC, Chang FR, Lin CY, et al. J Nat Prod. 2002;65:470. doi: 10.1021/np0105578. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Chen JW, Xu SS, et al. Bioorg Med Chem Lett. 2012;22:2717. doi: 10.1016/j.bmcl.2012.02.109. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Chen JW, Li X. J Nat Prod. 2011;74:2477. doi: 10.1021/np200708q. [DOI] [PubMed] [Google Scholar]
- 10.Oberlies NH, Chang CJ, McLaughlin JL. J Med Chem. 1997;40:2102. doi: 10.1021/jm9700169. [DOI] [PubMed] [Google Scholar]
- 11.Taylor L. Technical Data Report for Graviola (Annona muricata) Austin, TX: Sage Press Inc; 2005. [Google Scholar]
- 12.Rieser MJ, Gu ZM, Fang XP, Zeng L, Wood KV, McLaughlin JL. J Nat Prod. 1996;59:100. doi: 10.1021/np960037q. [DOI] [PubMed] [Google Scholar]
- 13.Chang FR, Liaw CC, Lin CY, Chou CJ, Chiu HF, Wu YC. Planta Med. 2003;69:241. doi: 10.1055/s-2003-38485. [DOI] [PubMed] [Google Scholar]
- 14.Torres MP, Rachagani S, Purohit V, et al. Cancer Lett. 2012;323:29. doi: 10.1016/j.canlet.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alali FQ, Liu XX, McLaughlin JL. J Nat Prod. 1999;62:504. doi: 10.1021/np980406d. [DOI] [PubMed] [Google Scholar]
- 16.Dai Y, Hogan S, Fau-Schmelz EM, et al. Nutr Cancer. 2011;63:795. doi: 10.1080/01635581.2011.563027. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez-Coloma A, Guadano A, de Ines C, Martinez-Diaz R, Cortes D. Z Naturforsch C: J Biosci. 2002;57:1028. doi: 10.1515/znc-2002-11-1213. [DOI] [PubMed] [Google Scholar]
- 18.Sun S, Liu J, Kadouh H, Sun X, Zhou K. Bioorg Med Chem Lett. 2014;24:2773. doi: 10.1016/j.bmcl.2014.03.099. [DOI] [PubMed] [Google Scholar]
- 19.Sun S, Liu J, Zhou N, Zhu W, Dou QP, Zhou K. Bioorg Med Chem Lett. 2015;26:4382. doi: 10.1016/j.bmcl.2015.06.038. [DOI] [PubMed] [Google Scholar]
- 20.McCloud TG, Smith DL, Chang CJ, Cassady JM. Experientia. 1987;43:947. doi: 10.1007/BF01951681. [DOI] [PubMed] [Google Scholar]
- 21.Rupprecht JK, Hui YH, McLaughlin JL. J Nat Prod. 1990;53:237. doi: 10.1021/np50068a001. [DOI] [PubMed] [Google Scholar]
- 22.Fang XP, Rieser MJ, Gu ZM, Zhao GX, McLaughlin JL. Phytochem Anal. 1993;4:49. [Google Scholar]
- 23.Fujimoto Y, Murasaki C, Shimada H, et al. Chem Pharm Bull. 1994;42:1175. [Google Scholar]
- 24.Gu ZM, Fang XP, Zeng L, Kozlowski JF, McLaughlin JL. Bioorg Med Chem Lett. 1994;4:473. [Google Scholar]
- 25.Shi G, Gu ZM, He K, et al. Bioorg Med Chem. 1996;4:1281. doi: 10.1016/0968-0896(96)00114-9. [DOI] [PubMed] [Google Scholar]
- 26.Wu FE, Gu ZM, Zeng L, et al. J Nat Prod. 1995;58:830. doi: 10.1021/np50120a002. [DOI] [PubMed] [Google Scholar]
- 27.Liaw CC, Yang YL, Chen M, et al. J Nat Prod. 2008;71:764. doi: 10.1021/np0704957. [DOI] [PubMed] [Google Scholar]
- 28.Born L, Lieb F, Lorentzen JP, et al. Planta Med. 1990;56:312. doi: 10.1055/s-2006-960967. [DOI] [PubMed] [Google Scholar]
- 29.Zeng L, Ye Q, Oberlies NH, et al. Nat Prod Rep. 1996;13:275. doi: 10.1039/np9961300275. [DOI] [PubMed] [Google Scholar]
- 30.Yuan S-SF, Chang H-L, Chen H-W, et al. Life Sci. 2003;72:2853. doi: 10.1016/s0024-3205(03)00190-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


