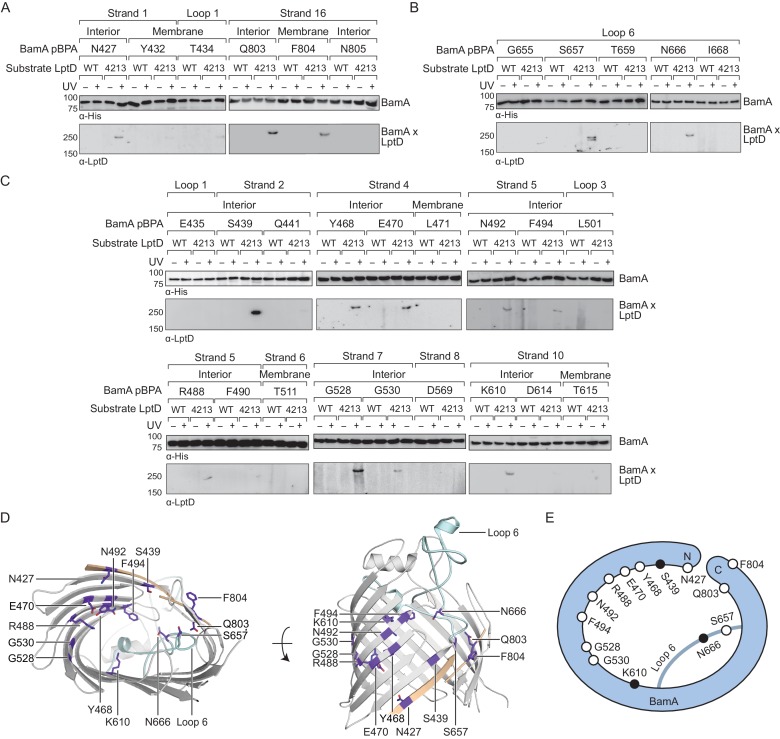
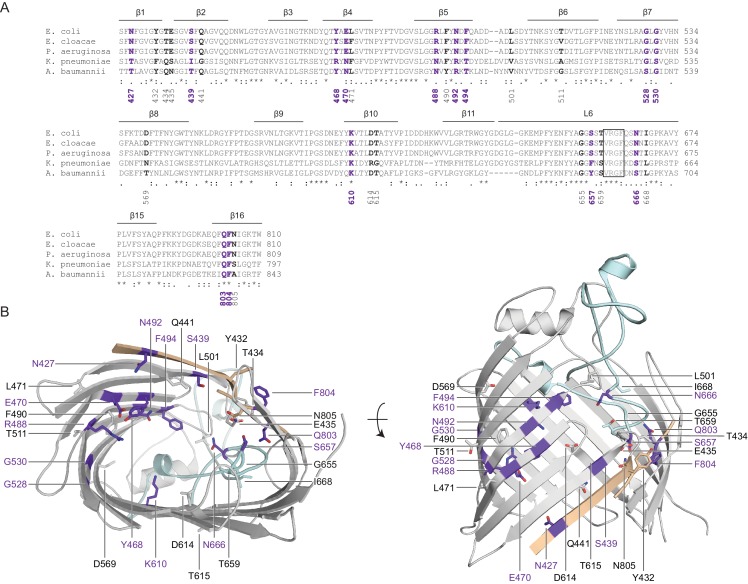
Figure 1. The interior wall and lateral gate of the BamA β-barrel form a substrate binding site.
(A–C) Residues at the lateral gate (A), L6 (B), and interior wall (C) of BamA interact with substrate LptD during assembly. MC4100 and lptD4213 (NR698) strains (expressing WT LptD or LptD4213, respectively) harboring the amber suppression system and expressing a His-tagged BamA (containing pBPA) were either left untreated or irradiated with UV light. Crosslinked adducts of BamA and substrate LptD/LptD4213 were identified by immunoblot analyses after Ni-NTA affinity purification. The orientation of the side chain of each residue in BamA substituted with pBPA is indicated (i.e., facing towards the membrane or interior of BamA). (D) Specific sites in the BamA β-barrel that interact with substrate LptD. Residues substituted with pBPA that crosslink to substrate are colored in purple. The first and last β-strands are colored in tan while the L6 loop is colored in cyan. Images were generated in PyMOL using the crystal structure of the BamA β-barrel from the E. coli BamABCDE complex (PDB: 5D0O). (E) Cartoon schematic of all sites in BamA that crosslink to substrate LptD. The view shown is the same as in the left panel of (D). Residues colored in black represent general substrate binding sites tested for crosslinking to full-length substrates (Figure 2). Additional views that include residues that do not form crosslinks are shown in Figure 1—figure supplement 1.