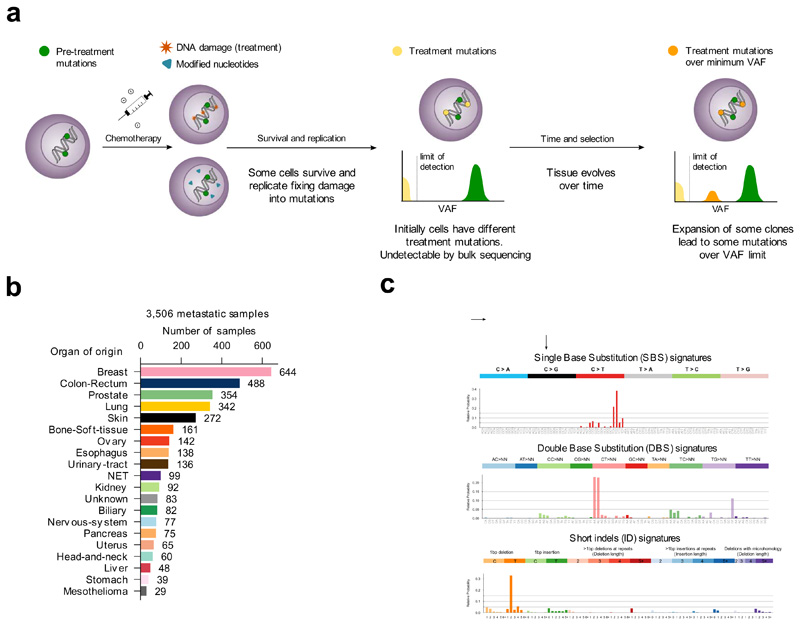
Figure 1. Mutational signatures active in metastatic tumors.
(a) Tumor cells bear mutations at the time of treatment contributed by different mutational processes. Some treatments directly damage the DNA, while others alter the pool of nucleotides, potentially causing the death of a large number of cells. Surviving cells harbor treatment-induced mutations caused by unrepaired DNA damage, the consequences of misincorporated nucleotide analogs or introduced by error-prone polymerases during repair. These treatment mutations are private to each surviving cell after the first round of replication, have low variant allele frequencies (VAF), and are undetectable through bulk sequencing. Pre-treatment mutations are present at higher VAF. Some surviving cells may grow faster than their neighbors to occupy the space opened by massive death of tumor cells. Over time, these faster-growing cells will undergo clonal expansion and their progeny will represent a larger fraction of the population, effectively amplifying their genetic material within the tumor pool. At the time of biopsy of the metastasis, the VAF of treatment mutations present in the original surviving cells may rise above the threshold of detection of bulk sequencing.
(b) Composition of the metastatic cohort in terms of organ of origin of the primary. The color code of organs of origin is used in subsequent figures. NET: Neuroendocrine tumors.
(c) Example SBS, DBS and ID signatures extracted from the metastatic cohort using SignatureAnalyzer. The profiles of all signatures identified using both methods appear in the Supplementary Note and Supplementary Datasets.