Figure 1.
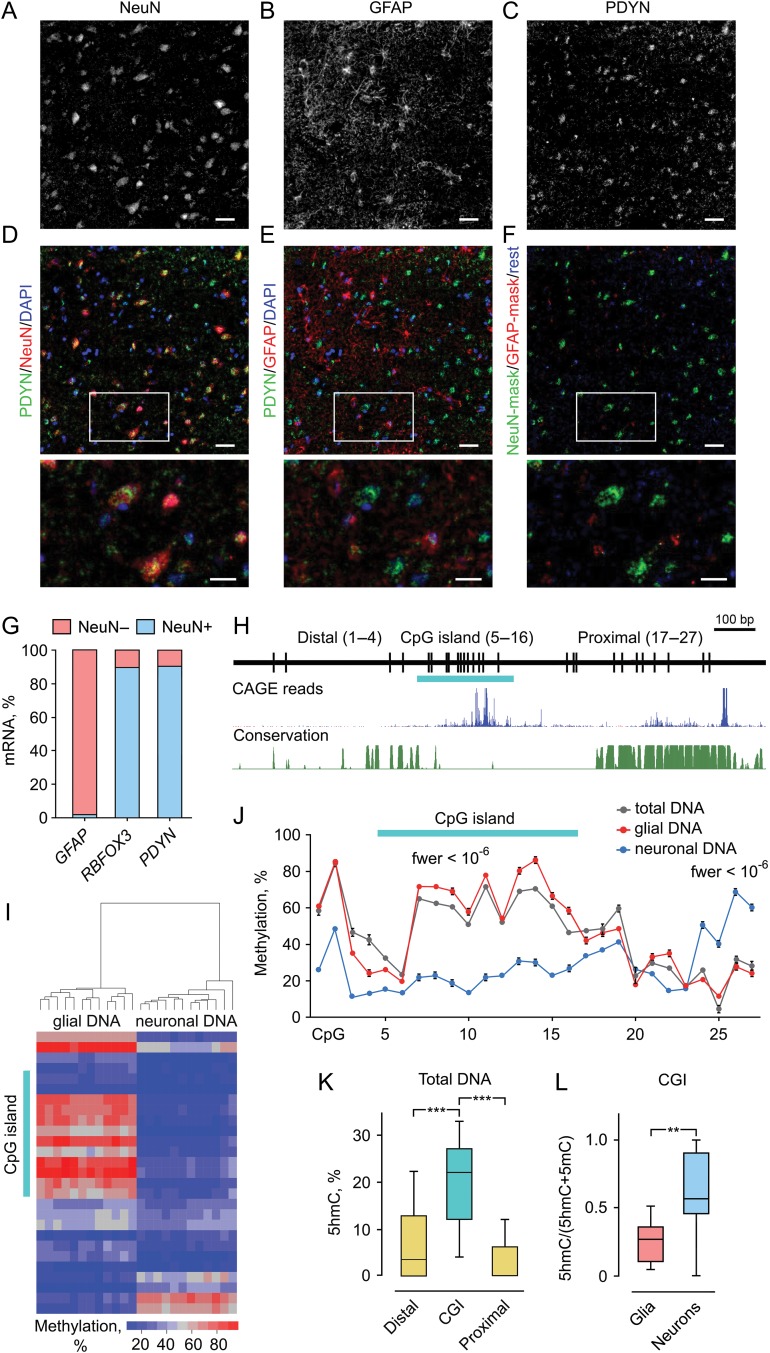
Neuronal expression of PDYN in human dlPFC is associated with 2 DMRs and elevated 5hmC levels in the promoter CGI. (A–F) Immunofluorescence micrographs showing the distribution of PDYN immunoreactivity (ir) in NeuN-ir neurons (A and D) and GFAP-ir astrocytes (B and E) in the cortex. PDYN-ir located in the cytosol and proximal processes of NeuN-ir neurons (C) was mainly co-localized with NeuN-ir (D). Minor PDYN-ir was co-localized with GFAP-ir (E) located in astrocyte cell bodies and processes (B). Neuronal PDYN-ir is shown in green, astrocyte PDYN-ir in red, and PDYN-ir not co-existing with GFAP-ir or NeuN-ir in deep blue (F). Lower row shows magnified images of the area outlined in (D–F). The majority of PDYN-ir pixels especially those with high intensity are found in neurons. Scale bars, 50 μm (A–F); 25 μm (magnified images in the lower row). (G) Levels of RBFOX3 (neuronal marker) and PDYN mRNAs are high in NeuN while those of GFAP (astrocyte marker) in non-NeuN. Bar graphs show average mRNA amount in the NeuN+ and NeuN− fractions as % of total amount of these mRNAs. Nuclei were isolated by FANS from dlPFC of 3 subjects, mRNA analyzed using ddPCR and normalized to total RNA. (H) Diagram depicts CpG positions, short CpG island (CGI) (horizontal bar), and Distal and Proximal regions in 1.42 kb human PDYN promoter. Genome browser shot shows TSSs in human PDYN promoter (FANTOM5 UCSC track, GRCh37/hg19), followed by phastCons track showing conserved elements. (I) Heatmap depicting methylation levels measured by pyrosequencing across PDYN promoter. Clusters of neuronal and non-neuronal DNA are distinguished based on methylation levels. (J) Two neuronal DMRs identified by “bumphunter”. DMR1 (CpGs 1–19) is hypo- while DMR2 (CpGs 24–27) hypermethylated in neurons. The CGI CpGs demonstrate largest and most significant differences among those in DMR1. Average methylation level for 12 subjects and the errors of the means are shown. fwer values for DMRs computed using bootstrap method, as detailed in the “Materials and Methods” section, are indicated. (K) The 5hmC levels are significantly higher in the CGI compared to the Distal or Proximal promoter regions in total tissue DNA analyzed by 5hmC-qAMP assay (n = 21 subjects). (L) A proportion of 5hmC assessed as the (5hmC)/(5hmC + 5mC) ratio in the CGI is significantly higher in neurons compared to glial cells analyzed by 5hmC-qAMP and qAMP assays (n = 12 subjects). The bar graphs show averages and the errors of the means. In box plots, center line is the median, box spans the interquartile range (IQR), and whiskers are 1.5 × IQR from box limits. **P < 0.01, ***P < 0.001. See also Supplementary Figures S1–S3.