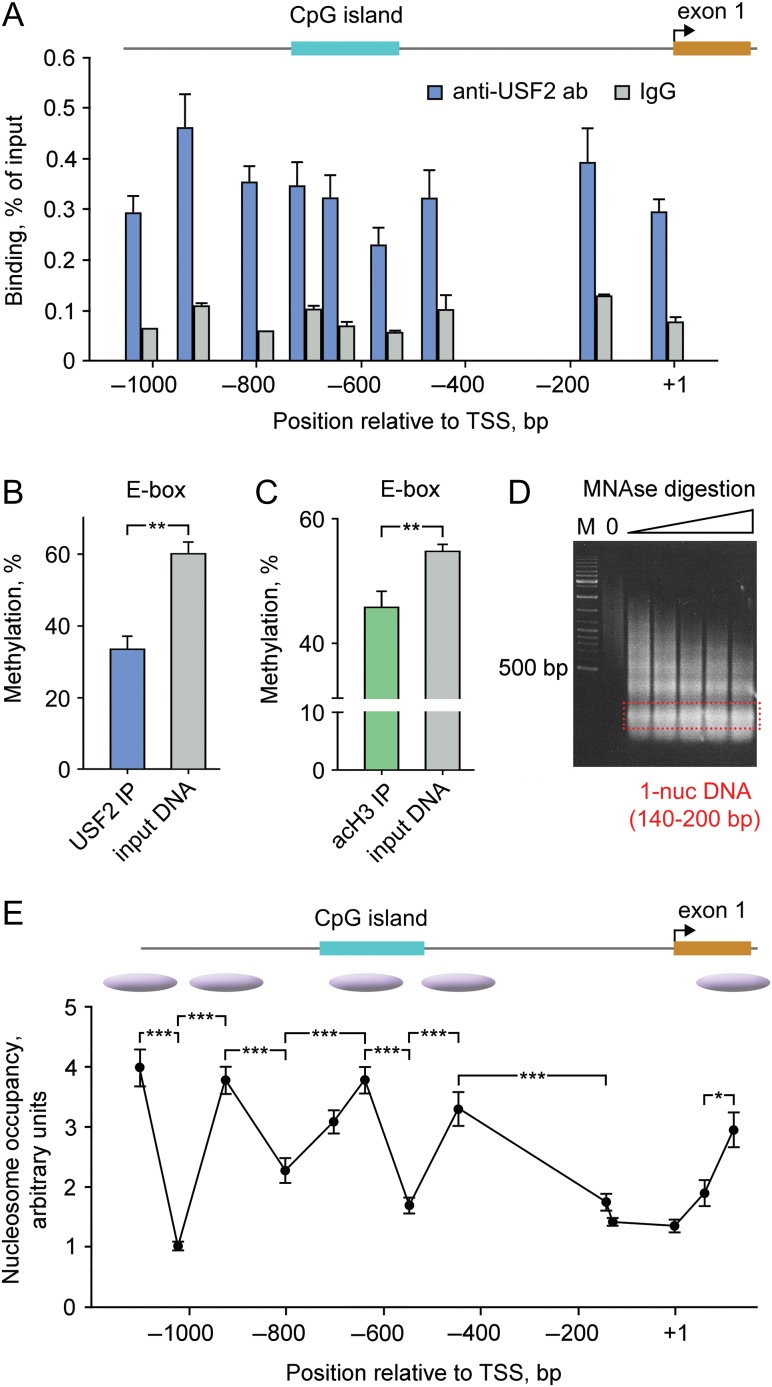
Figure 4.
USF2 binds to hypomethylated PDYN promoter in the human dlPFC. (A) Enrichment of USF2 across PDYN promoter. ChIP-qPCR results are shown as levels of DNA immunoprecipitated by anti-USF2 antibodies or control IgG for 9 amplicons. Six tissue samples all of each pooled from three subjects, altogether from 18 subjects were analyzed. Post hoc Tukey HSD-test showed significantly higher binding of USF2 comparing to control IgG (3.9-fold; P < 0.001) to the promoter. Bar graphs show mean ± SEM. (B and C) Methylation of the PDYN promoter E-box CpG 12 associated with USF2 (B) or acetylated histone 3 (H3K9/K14 acetylation, an activatory mark; (C)) was significantly lower compared to that of the total tissue DNA. Methylation was analyzed in DNA immunoprecipitated either with anti-USF2 antibodies or anti-H3-Ac antibodies by qAMP assay using PCR primer Set 7 (see Supplementary Table S3). Six tissue samples all of each pooled from 3 subjects, altogether from 18 subjects were analyzed. Bar graphs show mean ± SEM. **P < 0.01 (t-test). (D) Marker (M) and DNAs digested with increasing amounts of MNAse (1.25, 1.5, 1.75, 2.0, and 2.25 units) on an agarose gel. (E) qPCR analysis of MNAse-digested chromatin from dlPFC. Nucleosome occupancy and inferred positioning are shown. The bar graphs (A–C) and data points (E) show averages for 6 samples pooled from 18 subjects and the errors of the means. The lines connecting data points do not imply levels between points. Bent arrow, proximal TSS. *P < 0.05, **P < 0.01, ***P < 0.001. For experimental details, see Supplementary Figure S5.