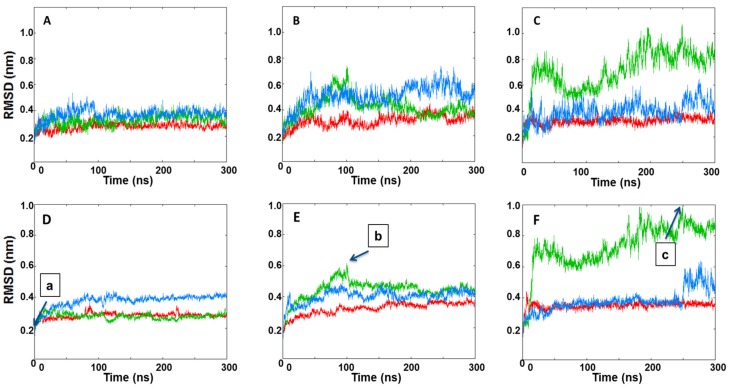
Figure 5.
The RMSDs of IRE1 face-to-face dimer Cα atoms during the three MD simulation replicas of (A) a native face-to-face crystal dimer structure (PDB code: 3P23), (B) KIRA docked in PDB 3P23 dimer, (C) protein–protein docked pose of PDB 4U6R in the face-to-face dimer form. Interface RMSDs of IRE1 face-to-face dimer Cα atoms during the three MD simulation replicas of (D) a native face-to-face crystal dimer structure (PDB code: 3P23), (E) KIRA docked in PDB 3P23 dimer, (F) protein–protein docked pose of PDB 4U6R in the face-to-face dimer form. Replicas 1, 2, and 3 are represented in red, green, and blue, respectively. Individual frames of the MD simulations labeled (a–c) are shown in Figure S4.