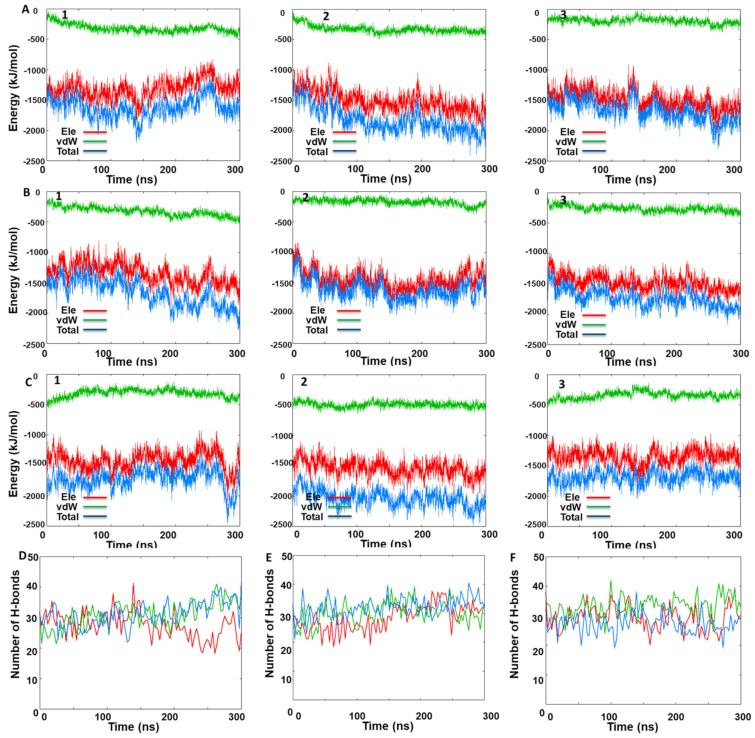
Figure 8.
IRE1 back-to-back dimer MD simulation data for the three MD replicas. Time-dependent interaction energy profiles for monomer A with monomer B during the MD simulations of (A) native back-to-back dimer (PDB code: 4YZC), (B) KIRA docked in the 4YZC structure, and (C) protein–protein docked pose of PDB 4U6R in the back-to-back dimer. Hydrogen bond analysis between the monomers A and B during the three MD replicas for (D) native back-to-back crystal dimer (PDB code: 4YZC), (E) KIRA docked in PDB 4YZC structure, and (F) protein–protein docked pose of PDB 4U6R in the back-to-back dimer form.