Figure 1.
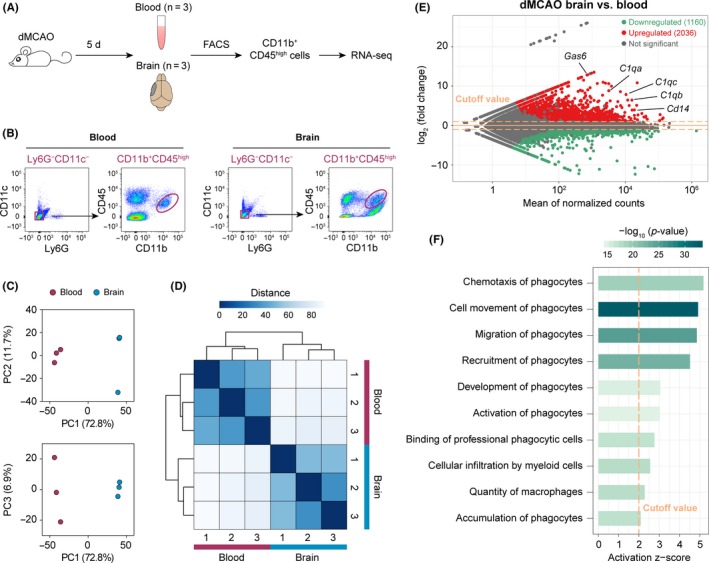
Whole‐genome transcriptomic profiling of monocytes/macrophages in the mouse blood and brain after ischemic stroke. A, Illustration of experimental design. Mice were subjected to permanent focal cerebral ischemia induced by distal middle cerebral artery occlusion (dMCAO). The monocyte/macrophage population (CD11b+CD45high cells) were sorted from blood and brain by FACS at 5 days after dMCAO and subjected to RNA‐seq. There were three biological replicates in each group. B, Representative FACS density plots illustrating the cells sorted for RNA‐seq. Cells were gated as Ly6G−CD11c−CD11b+CD45high. C, Principal component analysis was performed on RNA‐seq expression profiles of FACS‐sorted cells. Samples in the same experimental group clustered together on principal component 1 (PC1). D, Heatmap of the sample‐to‐sample Euclidean distances illustrating high similarities among the samples in the same group and disparate profiles among samples in different groups. E, MA plot of average RNA‐seq expression differences showing the differentially expressed genes (DEGs; fold change >2 or <−2, adjusted P‐value <.05) in monocytes/macrophages from the post‐dMCAO brain versus blood. F, Ingenuity Pathway Analysis (IPA) performed on the DEGs in cells from the brain versus blood predicted several functions of phagocytes to be significantly activated (activation z‐score >2, P‐value of overlap <.01)
