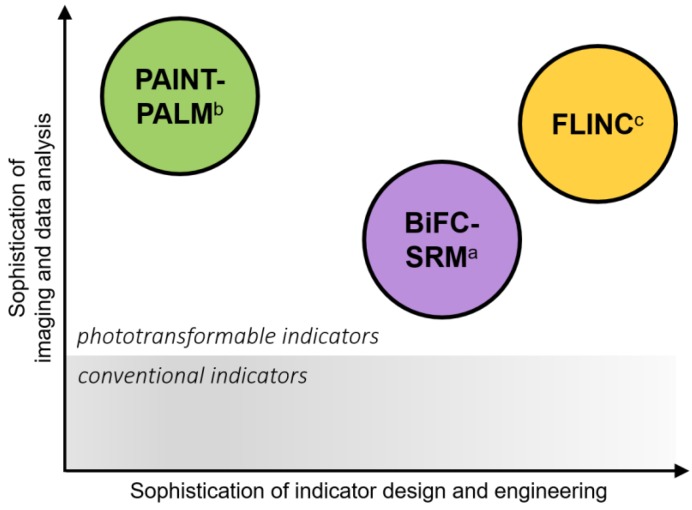
Figure 3.
Accessibility of genetically encoded indicators derived from phototransformable fluorescent proteins for functional super-resolution imaging. a: BiFC-based indicators for SRM (BiFC-SRM) borrow the design basics from conventional BiFC indicators. The main challenges for engineering these indicators are two-folds: The first is to identify split site(s) that permits reconstitution of fluorescence and phototransformation; The second is fast chromophore maturation after complementation. Methods for SRM are straightforwardly adopted from SOFI, PALM, or RESOLFT with little modification. b: PAINT-PALM indicators are simply engineered by fusing a phototransformable FP to the diffusive protein of interest. For SRM of biomolecule interactions, both camera exposure and single-molecule localization algorithm must be fine-tuned. The goal is to filter out unbound and motion-blurred emitters (fail if exposure is too short and inclusion criteria set too low); in the meantime, bound emitters must assume well-defined Gaussian PSFs to be successfully localized (fail if exposure is too long and inclusion criteria set too high). c: Design of FLINC indicator is generalizable to the detection of both biochemical activities and PPI. Main limitation is the incomplete mechanistic understanding of FLINC phenomenon, which limits the discovery of new FLINC protein pairs. To generate super-revolved map of biomolecular activities, cumulant values must be properly normalized to eliminate bias from uneven distribution of indicators across the cell.