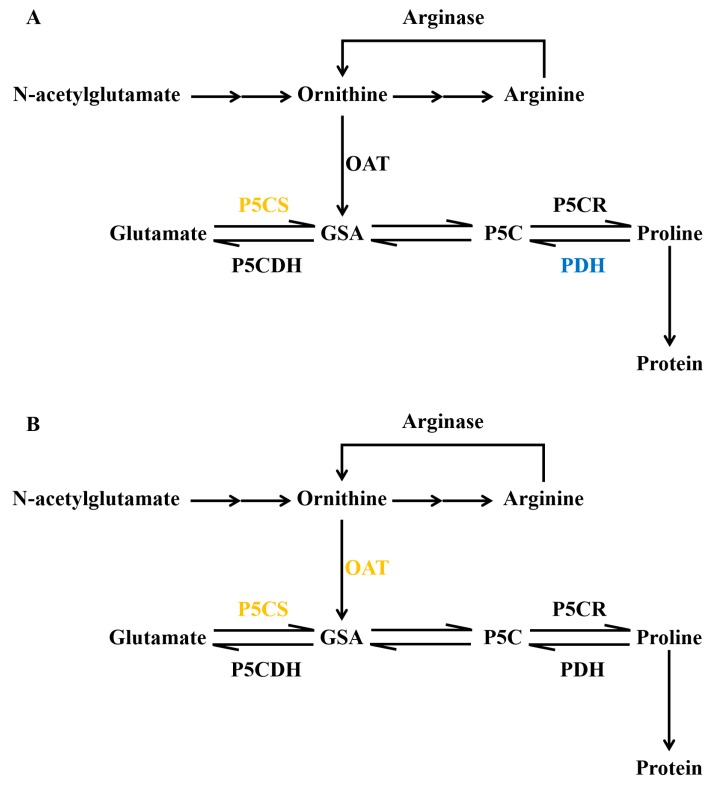
Figure 6.
Major DEGs involved in proline metabolism in B. napus. (A) Proline metabolism pathway DEGs in CK-R (CK, roots) vs. D-R (drought, roots) and CK-R vs. S-R (salinity, roots). (B) Proline metabolism pathway DEGs in CK-L (CK, leaves) vs. D-L (drought, leaves). P5CS (Δ1-pyrroline-5-carboxylate synthetase), P5CDH (pyrrolidine-5-carboxylic acid dehydrogenase), GSA (glutamate-γ-semialdehyde), P5C (pyrroline-5-carboxylic acid), P5CR (Δ1-pyrroline-5-carboxylate reductase), PDH (proline dehydrogenase), and OAT (ornithine aminotransferase) function in proline metabolism. Yellow indicates that the underlying DEGs were upregulated, while blue indicates that the underlying DEGs were downregulated.