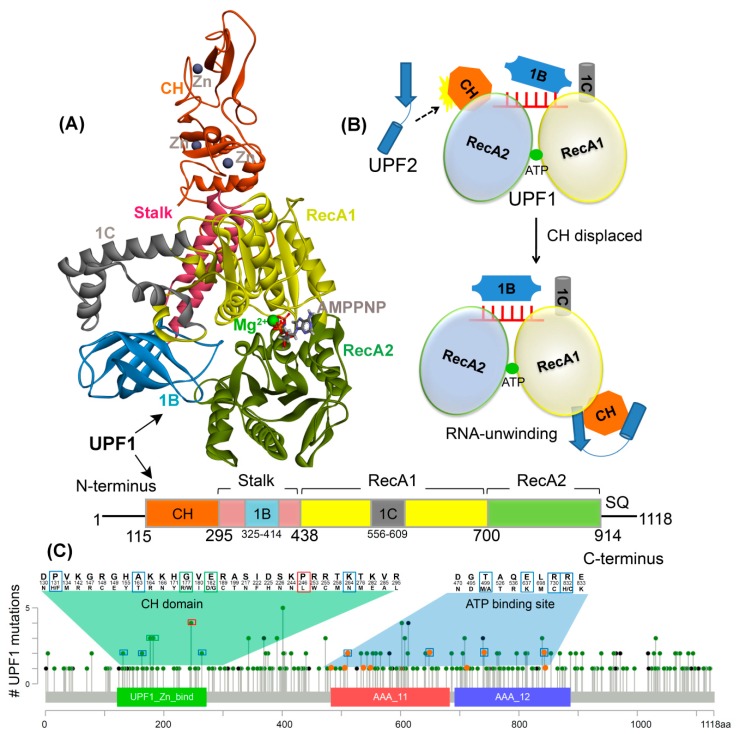
Figure 1.
UPF1 structure and mutations from different cancer types. (A) Crystal structure of UPF1 (PDB: 2WJY [10]) representing different domains, ATP analogue (i.e., AMPPNP), and Zn/Mg2+ ions. (B) Schematic model of UPF1 activation mechanism showing how the RNA binding and ATPase/unwinding activity are modulated by UPF2 in an NMD cycle. In the absence of UPF2, the UPF1 has extended RNA interactions via the RecA1, RecA2, 1B, and 1C domains and low RNA-unwinding properties. The CH-domain is displaced upon UPF2 binding (black dotted arrow) and domain 1B changes to a conformation where it does not clamp on the RNA 3′ end and as a result, the RNA-unwinding activity is increased [11]. (C) UPF1 gene mutated in cancer, data retrieved from the cBioPortal database [29]. The frequency of a residue mutation equal to 2, 3, and 4 are marked in blue, green, and red boxes respectively. The mutations from the ATP-binding site are shown in orange. Color scheme: Zn and carbon in grey, Mg in green, oxygen in red, hydrogen in silver, nitrogen in blue, and sulfur in orange.