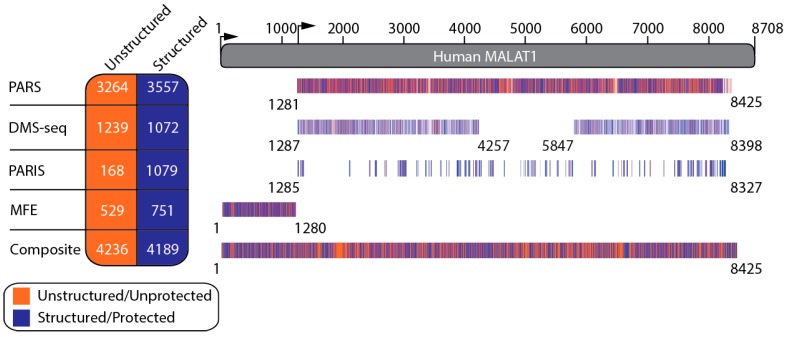
Figure 1.
Overview of structural assignments for each nucleotide in human MALAT1. PARS data [21], DMS-seq data [22], PARIS data [26,27], and minimum free energy (MFE) calculations [29,32] were collectively used to construct a composite structural landscape for each nucleotide in MALAT1 (nts 1–8425). The orange lines represent individual nucleotide positions that were designated as unstructured based on PARS datasets and MFE calculations and unprotected adenosine or cytidine residues based on the DMS-seq dataset. The blue bars represent individual nucleotide positions that were designated as structured based on PARS datasets and MFE calculations and as protected adenosine or cytidine residues based on the DMS-seq dataset. The number of nucleotides that are unstructured/unprotected (orange column) or structured/protected (blue column) for PARS, DMS-seq, PARIS, MFE, and the composite model are listed in the table to the left. PARIS data that overlap with unstructured or structured regions of the MALAT1 transcript are presented as either orange or blue, respectively. The short arrow represents the transcription start site of the full-length malat1 gene (nts 1–8708), while the tall arrow represents the transcription start site of the major variant of the malat1 gene (nts 1284–8708).