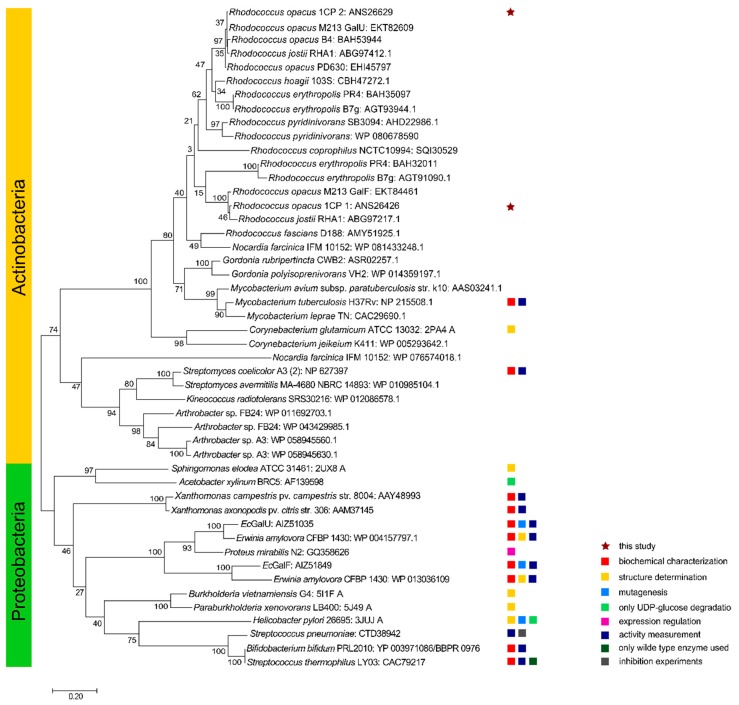
Figure 2.
Phylogenetic tree of RoGalU1 and RoGalU2 with similar characterized or related UDP-glucose pyrophosphorylase sequences. The evolutionary history was inferred by using the Maximum Likelihood method and JTT matrix-based model [49]. The tree with the highest log likelihood (−13775.30) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measuring the number of substitutions per site. This analysis involved 48 amino acid sequences. There was a total of 375 positions in the final dataset. Evolutionary analyses were conducted in MEGA X [47].