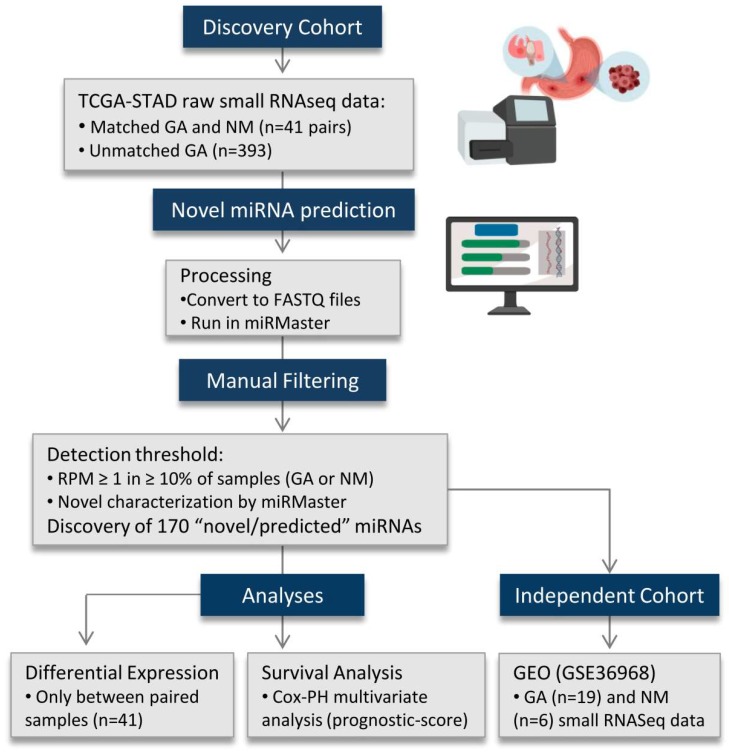
Figure 3.
Flow diagram outlining the major steps of the study. Novel miRNAs were predicted using the miRMaster tool on small RNA sequencing data from the TCGA stomach cohort. One-hundred and seventy novel miRNA candidates were curated based on their expression in non-malignant and malignant samples. These sequences showed a global overexpression in gastric tumors when compared to non-malignant samples. A second, publicly available, small RNA sequencing cohort (GSE36968: 19 tumors and six non-malignant gastric samples) was assessed. Forty-three novel miRNA candidates were detected in both the discovery and independent cohorts. The novel miRNAs were submitted to differential expression and overall survival analyses (univariate and multivariate analyses). GA: Gastric adenocarcinoma; NM: Non-malignant; RPM: Reads per million; COX-PH: Cox proportional-hazards.