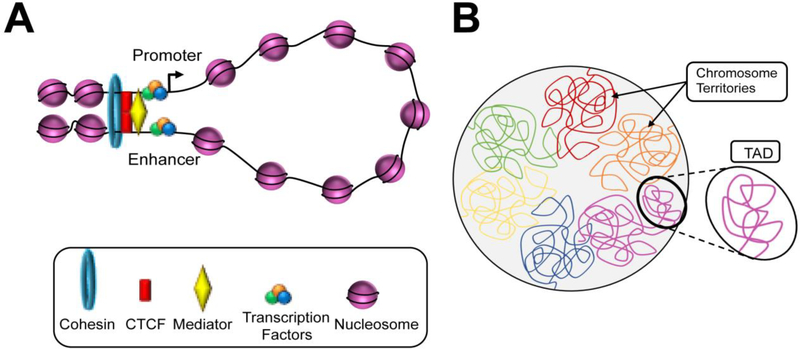
Figure 2. Three-dimensional chromosome organization.
(A) Locally, at the submegabase scale, chromatin loop brings the distant cis-regulartory elements (e.g. enhancers) into close spatial proximity with its target promoter to regulate transcription. Architectural proteins and complexes including CTCF, mediator, and cohesin facilitate the formation of DNA loops. (B) On a larger scale, multiple loci undergoing regulatory interactions are grouped into topologically associated domains (TADs). The sequences within the same TADs have a high degree of interaction, which correlates with coordinated gene expression. Furthermore, all TADs are assembled into two spatial compartments (not shown). Lastly, each chromosome tends to locate in a discrete area within the nucleus, namely chromosome territories