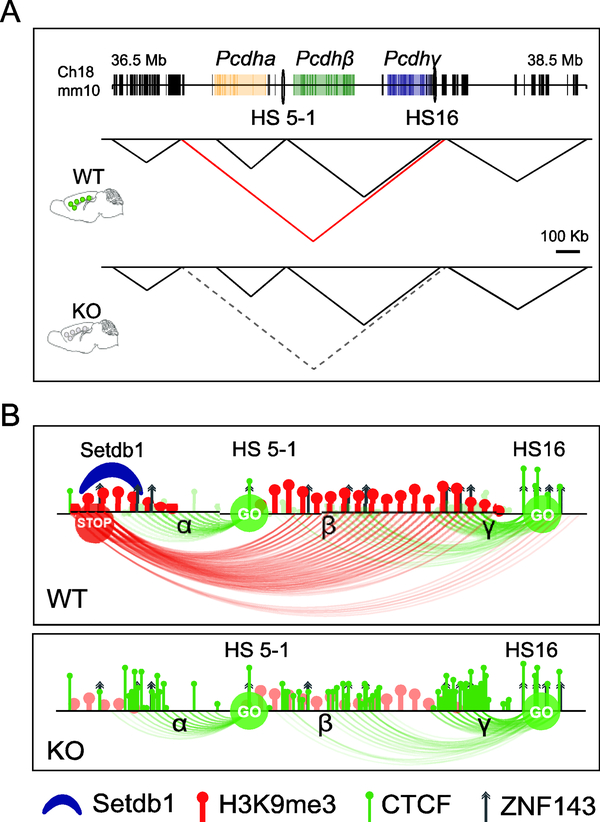
Figure 1: Selective loss of a large megabase TAD at the clustered Protocadherin gene locus after neuronal ablation of Kmt1e/Setdb1 methyltransferase.
(A) Schematic overview of TAD landscape of adult cortical neurons in 2Mb portion of chromosome 18, shown for wildtype (WT) and conditional Setdb1 knock-out (KO), as indicated. Dotted triangle in KO (corresponding to red triangle in WT) demarcates TAD that is massively weakened in KO neurons. Notice that subTADs and surrounding TAD landscape is minimally affected. (B) Chromatin structure and function at the clustered Protocadherin locus. Schematic presentation of 1 megabase TAD at site of Protocadherin genes (see A), illustrating repressive chromatin (‘STOP’) in TAD periphery connecting to intraTAD enhancer sequences (‘GO’). Top, WT, Bottom, KO. Loss of SETDB1 in neurons results in histone hypomethylation, massive excess of CTCF across the TAD, loss of repressive loopings at enhancers and zinc finger protein (incl. ZNF143) binding sites, triggering excessive transcription. See ref. [42] for details.