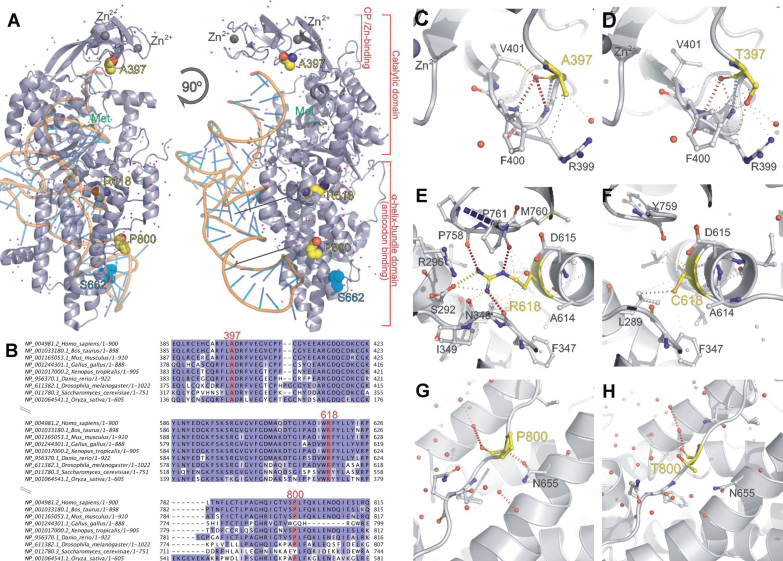
Fig.1.
Structural studies of CMT-associated missense variants. (A) Structure of the human MARS protein (PDB ID 5goy) complexed with methionine (green) and docked with a tRNA molecule. Residues Ala 397, Arg 618 and Pro 800, found mutated in CMT affected individuals, are shown in yellow spheres. (B) Sequence alignment showing conservation of residues 397, 618 and 800. Internal contacts calculated using the software Arpeggio for each residue are shown: (C) A397, (D) T397, (E) R618, (F) C618, (G) P800, (H) T800. Contacts in C-H are shown in dotted lines: black lines indicate hydrophobic interactions (short distance, <5 Å), red show hydrogen bonds, yellow polar interactions and grey carbon-π-aromatic interactions. The catalytic domain of MARS, as well as of other class I aminoacyl-tRNA synthetases, is composed by a Rossmann fold domain and a connective polypeptide (CP) domain formed by β ribbon with two zinc fingers (A) [20]. The tRNA binding is established in an all-helical domain in the C-terminal portion of the protein (A). An extrapolation of the predicted tRNA binding position is shown (A), based on superimposition with tRNA-bound Ile synthetase (1FFY).