Fig. 4.
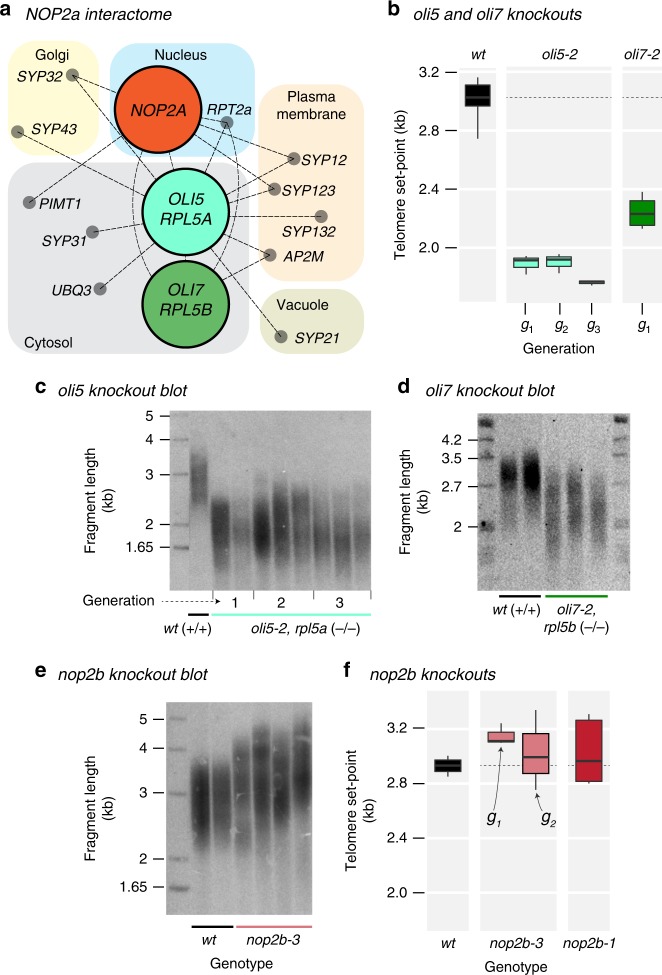
Specific components of ribosome assembly and rRNA biogenesis pathway control the establishment of telomere length set point in Arabidopsis. a Protein interactions (‘interactome’) of OLI2/NOP2A, OLI5/RPL5A, and OLI7/RPL5B, genes previously identified in a forward genetic screen for OLIGOCELLULA cell proliferation mutants29, are shown. The edges in the network connect predicted interacting proteins (nodes), which are grouped by their predicted cellular locations. b Boxplots indicate the significantly shorter telomeres in rpl5a/oli5-2 and rpl5b/oli7-2 mutants relative to the wild type. c, d Examples of TRF southern blots of rpl5a/oli5-2 and rpl5b/oli7-2 mutants. Arabidopsis RPL5A/OLI5 (c) and RPL5B/OLI7 (d) T-DNA knockout plants harbor stable telomere phenotypes with a new shorter length set point. e Example of TRF southern blot for nop2b-3 mutants. Unlike NOP2A mutants, Arabidopsis NOP2B T-DNA knockout plants harbor telomere length in the wild type range. f Boxplots (midline = median, box = interquartile range (IQR), whiskers = 1.5*IQR) indicate telomere lengths in nop2b-1 and nop2b-3 mutants relative to the wild type. The y-axis scale has been extended to facilitate comparisons with the minimum TRF of rpl5a/oli5 and rpl5b/oli7 in b.