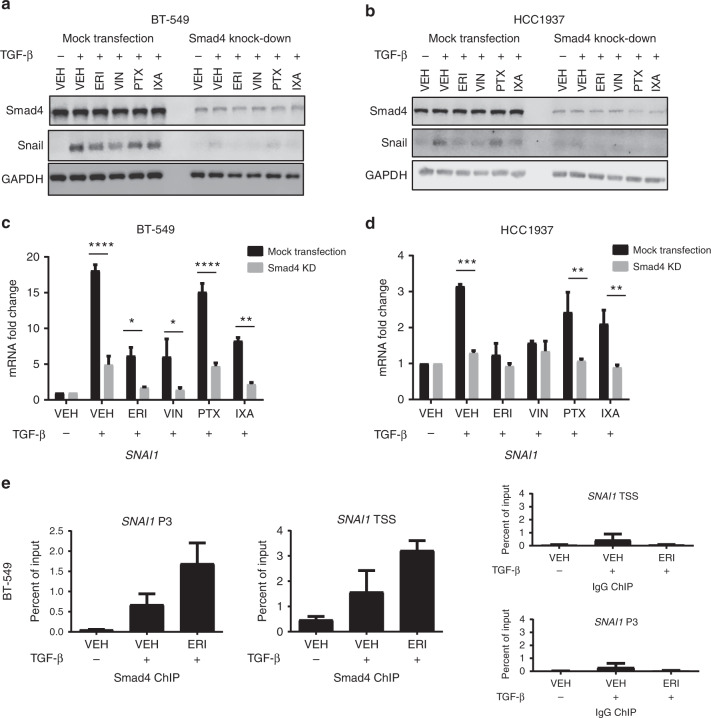
Fig. 3.
Smad4 is required for Snail expression. a, b Smad4 was knocked-down in BT-549 and HCC1937 cells by siRNA after which cells were serum-starved for 18 h, pre-treated with microtubule targeting agents for 2 h, and stimulated with TGF-β for 3 h. Cell lysates were subject to immunoblotting and probed for Smad4, Snail, and GAPDH. c, d qRT-PCR analysis of SNAI1 transcript after siRNA-mediated knock-down of Smad4. Cells were serum-starved for 18 h, pre-treated with microtubule targeting agents for 2 h, and stimulated with TGF-β for 1 h (BT-549) or 2 h (HCC1937). Data are the average of two independent experiments (±SEM). Statistical significance of Smad4 knock-down for each condition was determined using a two-way ANOVA with Holm-Sidak’s multiple comparison test. (****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05) e ChIP-qPCR of two distinct SNAI1 promoter regions. After 18 h of serum-starvation, BT-549 cells were pre-treated with either vehicle or eribulin for 2 h followed by stimulation with TGF-β for 1 h. Chromatin immunoprecipitation of SNAI1 P3 and SNAI1 TSS promoter sequences with Smad4 or control IgG are shown. Data are the mean ± SEM (N = 2) and reported as percentage of input chromatin for each sequence