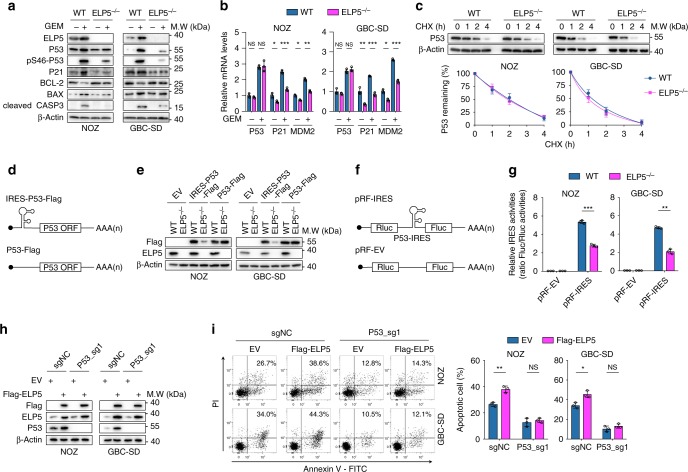
Fig. 5.
ELP5 promotes P53 expression by IRES-dependent translation. a ELP5 depletion substantially rescued the accumulation and activation of P53 and P53-mediated apoptosis under GEM treatment at IC50 for 72 h in GBC cells. b RT-qPCR confirmed that ELP5 depletion did not affect the P53 mRNA level with or without GEM treatment, but P53 target genes (i.e., P21 and MDM2) displayed dramatically reduced mRNA levels in ELP5−/− cells. c ELP5 depletion could not affect the P53 degradation rate in GBC cells. The P53 protein level was normalized to that of β-Actin, and the P53 protein levels in each cell were set as 1 at the 0 time point. d Schematic drawing of the exogenously delivered P53 construct with or without an IRES sequence within the 5′-UTR of the P53 ORF sequence, IRES-P53-Flag, and P53-Flag, respectively. UTR, untranslated region; ORF, open-reading frame. e The IRES-P53-Flag construct was inefficiently expressed in ELP5-depleted cells, but P53-Flag construct was adequately expressed in ELP5-depeleted and WT cells. f Schematic drawing of the bicistronic construct inserted with P53 IRES between Rluc and Fluc. Rluc, Renilla luciferase; Fluc, Firefly luciferase. g P53 IRES activity was notably reduced in ELP5-depleted cells, as assessed by the ratio of Fluc to Rluc via dual-luciferase reporter assay. h, i Ectopically expressed ELP5 in P53-knockout cells (h) could not rescue the reduced GEM-induced apoptosis caused by P53 depletion (i). Data represent the mean ± S.D., n = 3 independent experiments in b, c, g, i, error bars represent S.D. Unpaired Student’s t tests were used in b, g, i (NS, non-significant, *P < 0.05, **P < 0.01 and ***P < 0.001).